#9846 closed enhancement (fixed)
Allow fetching fit PDB models together with EMDB maps
| Reported by: | Owned by: | Tom Goddard | |
|---|---|---|---|
| Priority: | normal | Milestone: | |
| Component: | Volume Data | Version: | |
| Keywords: | Cc: | Elaine Meng | |
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
The following bug report has been submitted:
Platform: macOS-13.5.2-x86_64-i386-64bit
ChimeraX Version: 1.7.dev202309121847 (2023-09-12 18:47:21 UTC)
Description
Would like to be able to fetch EMDB maps together with fit PDB models. Oliver Clarke asked about this on the ChimeraX mailing list today.
Log:
Could not find tool "Tabbed Toolbar"
UCSF ChimeraX version: 1.7.dev202309121847 (2023-09-12)
© 2016-2023 Regents of the University of California. All rights reserved.
How to cite UCSF ChimeraX
> open 34838 fromDatabase emdb
Summary of feedback from opening 34838 fetched from emdb
---
note | Fetching compressed map 34838 from
https://files.wwpdb.org/pub/emdb/structures/EMD-34838/map/emd_34838.map.gz
Opened emdb 34838 as #1, grid size 256,256,256, pixel 1.08, shown at level
0.0224, step 1, values float32
> volume #1 level 0.08246
> open 8hju
Summary of feedback from opening 8hju fetched from pdb
---
note | Fetching compressed mmCIF 8hju from
http://files.rcsb.org/download/8hju.cif
8hju title:
Cryo-EM structure of native RC-LH complex from Roseiflexus castenholzii at
10,000 lux [more info...]
Chain information for 8hju #2
---
Chain | Description | UniProt
0 2 4 6 8 B E G I K O Q S U W | Beta subunit of light-harvesting 1 |
Q83XD2_9CHLR 1-55
1 3 5 7 9 A D F H J N P R T V | Alpha subunit of light-harvesting 1 |
Q83XD1_9CHLR 1-42
C | MULTIHEME_CYTC DOMAIN-CONTAINING PROTEIN | A7NQE7_ROSCS 1-320
L | Reaction center protein L chain | A7NQE8_ROSCS 1-315
M | Reaction center protein M chain | A7NQE8_ROSCS 335-641
X | SUBUNIT X |
Y | SUBUNIT Y |
Z | SUBUNIT Z |
Non-standard residues in 8hju #2
---
BCL — bacteriochlorophyll A
BPH — bacteriopheophytin A
DGA — diacyl glycerol
FE — Fe (III) ion
HEM — protoporphyrin IX containing Fe (HEME)
KGD — beta,psi-caroten-4-one (Keto-gamma-carotene)
MQE —
2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1
4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione (Menaquinone 11)
PGV —
(1R)-2-{[{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(palmitoyloxy)methyl]ethyl
(11E)-octadec-11-enoate (phosphatidylglycerol; 2-vaccenoyl-1-palmitoyl-Sn-
glycerol-3-phosphoglycerol)
> volume #1 level 0.0318
> style stick
Changed 23917 atom styles
> lighting soft
OpenGL version: 4.1 ATI-4.14.1
OpenGL renderer: AMD Radeon Pro 580 OpenGL Engine
OpenGL vendor: ATI Technologies Inc.
Python: 3.11.2
Locale: UTF-8
Qt version: PyQt6 6.3.1, Qt 6.3.1
Qt runtime version: 6.3.2
Qt platform: cocoa
Hardware:
Hardware Overview:
Model Name: iMac
Model Identifier: iMac18,3
Processor Name: Quad-Core Intel Core i7
Processor Speed: 4.2 GHz
Number of Processors: 1
Total Number of Cores: 4
L2 Cache (per Core): 256 KB
L3 Cache: 8 MB
Hyper-Threading Technology: Enabled
Memory: 32 GB
System Firmware Version: 515.0.0.0.0
OS Loader Version: 577.140.2~15
SMC Version (system): 2.41f2
Software:
System Software Overview:
System Version: macOS 13.5.2 (22G91)
Kernel Version: Darwin 22.6.0
Time since boot: 17 days, 22 hours, 43 minutes
Graphics/Displays:
Radeon Pro 580:
Chipset Model: Radeon Pro 580
Type: GPU
Bus: PCIe
PCIe Lane Width: x16
VRAM (Total): 8 GB
Vendor: AMD (0x1002)
Device ID: 0x67df
Revision ID: 0x00c0
ROM Revision: 113-D000AA-931
VBIOS Version: 113-D0001A1X-025
EFI Driver Version: 01.00.931
Metal Support: Metal 2
Displays:
iMac:
Display Type: Built-In Retina LCD
Resolution: Retina 5K (5120 x 2880)
Framebuffer Depth: 30-Bit Color (ARGB2101010)
Main Display: Yes
Mirror: Off
Online: Yes
Automatically Adjust Brightness: Yes
Connection Type: Internal
Installed Packages:
alabaster: 0.7.13
appdirs: 1.4.4
appnope: 0.1.3
asttokens: 2.2.1
Babel: 2.12.1
backcall: 0.2.0
beautifulsoup4: 4.11.2
blockdiag: 3.0.0
blosc2: 2.0.0
build: 0.10.0
certifi: 2022.12.7
cftime: 1.6.2
charset-normalizer: 3.2.0
ChimeraX-AddCharge: 1.5.11
ChimeraX-AddH: 2.2.5
ChimeraX-AlignmentAlgorithms: 2.0.1
ChimeraX-AlignmentHdrs: 3.4
ChimeraX-AlignmentMatrices: 2.1
ChimeraX-Alignments: 2.9.3
ChimeraX-AlphaFold: 1.0
ChimeraX-AltlocExplorer: 1.1.1
ChimeraX-AmberInfo: 1.0
ChimeraX-Arrays: 1.1
ChimeraX-Atomic: 1.47.2
ChimeraX-AtomicLibrary: 10.0.8
ChimeraX-AtomSearch: 2.0.1
ChimeraX-AxesPlanes: 2.3.2
ChimeraX-BasicActions: 1.1.2
ChimeraX-BILD: 1.0
ChimeraX-BlastProtein: 2.1.2
ChimeraX-BondRot: 2.0.4
ChimeraX-BugReporter: 1.0.1
ChimeraX-BuildStructure: 2.10.4
ChimeraX-Bumps: 1.0
ChimeraX-BundleBuilder: 1.2.2
ChimeraX-ButtonPanel: 1.0.1
ChimeraX-CageBuilder: 1.0.1
ChimeraX-CellPack: 1.0
ChimeraX-Centroids: 1.3.2
ChimeraX-ChangeChains: 1.0.2
ChimeraX-CheckWaters: 1.3.1
ChimeraX-ChemGroup: 2.0.1
ChimeraX-Clashes: 2.2.4
ChimeraX-ColorActions: 1.0.3
ChimeraX-ColorGlobe: 1.0
ChimeraX-ColorKey: 1.5.3
ChimeraX-CommandLine: 1.2.5
ChimeraX-ConnectStructure: 2.0.1
ChimeraX-Contacts: 1.0.1
ChimeraX-Core: 1.7.dev202309121847
ChimeraX-CoreFormats: 1.1
ChimeraX-coulombic: 1.4.2
ChimeraX-Crosslinks: 1.0
ChimeraX-Crystal: 1.0
ChimeraX-CrystalContacts: 1.0.1
ChimeraX-DataFormats: 1.2.3
ChimeraX-Dicom: 1.2
ChimeraX-DistMonitor: 1.4
ChimeraX-DockPrep: 1.1.2
ChimeraX-Dssp: 2.0
ChimeraX-EMDB-SFF: 1.0
ChimeraX-ESMFold: 1.0
ChimeraX-FileHistory: 1.0.1
ChimeraX-FunctionKey: 1.0.1
ChimeraX-Geometry: 1.3
ChimeraX-gltf: 1.0
ChimeraX-Graphics: 1.1.1
ChimeraX-Hbonds: 2.4
ChimeraX-Help: 1.2.1
ChimeraX-HKCage: 1.3
ChimeraX-IHM: 1.1
ChimeraX-ImageFormats: 1.2
ChimeraX-IMOD: 1.0
ChimeraX-IO: 1.0.1
ChimeraX-ItemsInspection: 1.0.1
ChimeraX-IUPAC: 1.0
ChimeraX-Label: 1.1.7
ChimeraX-ListInfo: 1.2
ChimeraX-Log: 1.1.5
ChimeraX-LookingGlass: 1.1
ChimeraX-Maestro: 1.8.2
ChimeraX-Map: 1.1.4
ChimeraX-MapData: 2.0
ChimeraX-MapEraser: 1.0.1
ChimeraX-MapFilter: 2.0.1
ChimeraX-MapFit: 2.0
ChimeraX-MapSeries: 2.1.1
ChimeraX-Markers: 1.0.1
ChimeraX-Mask: 1.0.2
ChimeraX-MatchMaker: 2.1.1
ChimeraX-MCopy: 1.0
ChimeraX-MDcrds: 2.6
ChimeraX-MedicalToolbar: 1.0.2
ChimeraX-Meeting: 1.0.1
ChimeraX-MLP: 1.1.1
ChimeraX-mmCIF: 2.12
ChimeraX-MMTF: 2.2
ChimeraX-Modeller: 1.5.9
ChimeraX-ModelPanel: 1.4
ChimeraX-ModelSeries: 1.0.1
ChimeraX-Mol2: 2.0
ChimeraX-Mole: 1.0
ChimeraX-Morph: 1.0.2
ChimeraX-MouseModes: 1.2
ChimeraX-Movie: 1.0
ChimeraX-Neuron: 1.0
ChimeraX-Nifti: 1.1
ChimeraX-NIHPresets: 1.1.9
ChimeraX-NRRD: 1.1
ChimeraX-Nucleotides: 2.0.3
ChimeraX-OpenCommand: 1.10.2
ChimeraX-PDB: 2.7.2
ChimeraX-PDBBio: 1.0.1
ChimeraX-PDBLibrary: 1.0.2
ChimeraX-PDBMatrices: 1.0
ChimeraX-PickBlobs: 1.0.1
ChimeraX-Positions: 1.0
ChimeraX-PresetMgr: 1.1
ChimeraX-PubChem: 2.1
ChimeraX-ReadPbonds: 1.0.1
ChimeraX-Registration: 1.1.1
ChimeraX-RemoteControl: 1.0
ChimeraX-RenderByAttr: 1.1
ChimeraX-RenumberResidues: 1.1
ChimeraX-ResidueFit: 1.0.1
ChimeraX-RestServer: 1.2
ChimeraX-RNALayout: 1.0
ChimeraX-RotamerLibMgr: 3.0
ChimeraX-RotamerLibsDunbrack: 2.0
ChimeraX-RotamerLibsDynameomics: 2.0
ChimeraX-RotamerLibsRichardson: 2.0
ChimeraX-SaveCommand: 1.5.1
ChimeraX-SchemeMgr: 1.0
ChimeraX-SDF: 2.0.1
ChimeraX-Segger: 1.0
ChimeraX-Segment: 1.0.1
ChimeraX-SelInspector: 1.0
ChimeraX-SeqView: 2.9
ChimeraX-Shape: 1.0.1
ChimeraX-Shell: 1.0.1
ChimeraX-Shortcuts: 1.1.1
ChimeraX-ShowSequences: 1.0.1
ChimeraX-SideView: 1.0.1
ChimeraX-Smiles: 2.1.2
ChimeraX-SmoothLines: 1.0
ChimeraX-SpaceNavigator: 1.0
ChimeraX-StdCommands: 1.12.2
ChimeraX-STL: 1.0.1
ChimeraX-Storm: 1.0
ChimeraX-StructMeasure: 1.1.2
ChimeraX-Struts: 1.0.1
ChimeraX-Surface: 1.0.1
ChimeraX-SwapAA: 2.0.1
ChimeraX-SwapRes: 2.2.2
ChimeraX-TapeMeasure: 1.0
ChimeraX-Test: 1.0
ChimeraX-Toolbar: 1.1.2
ChimeraX-ToolshedUtils: 1.2.4
ChimeraX-Topography: 1.0
ChimeraX-ToQuest: 1.0
ChimeraX-Tug: 1.0.1
ChimeraX-UI: 1.31.2
ChimeraX-uniprot: 2.3
ChimeraX-UnitCell: 1.0.1
ChimeraX-ViewDockX: 1.2.2
ChimeraX-VIPERdb: 1.0
ChimeraX-Vive: 1.1
ChimeraX-VolumeMenu: 1.0.1
ChimeraX-VTK: 1.0
ChimeraX-WavefrontOBJ: 1.0
ChimeraX-WebCam: 1.0.2
ChimeraX-WebServices: 1.1.1
ChimeraX-Zone: 1.0.1
colorama: 0.4.6
comm: 0.1.3
contourpy: 1.1.0
cxservices: 1.2.2
cycler: 0.11.0
Cython: 0.29.33
debugpy: 1.6.7
decorator: 5.1.1
docutils: 0.19
executing: 1.2.0
filelock: 3.9.0
fonttools: 4.41.1
funcparserlib: 1.0.1
grako: 3.16.5
h5py: 3.9.0
html2text: 2020.1.16
idna: 3.4
ihm: 0.38
imagecodecs: 2023.7.10
imagesize: 1.4.1
ipykernel: 6.23.2
ipython: 8.14.0
ipython-genutils: 0.2.0
ipywidgets: 8.0.7
jedi: 0.18.2
Jinja2: 3.1.2
jupyter-client: 8.2.0
jupyter-core: 5.3.1
jupyterlab-widgets: 3.0.8
kiwisolver: 1.4.4
line-profiler: 4.0.2
lxml: 4.9.2
lz4: 4.3.2
MarkupSafe: 2.1.3
matplotlib: 3.7.2
matplotlib-inline: 0.1.6
msgpack: 1.0.4
nest-asyncio: 1.5.6
netCDF4: 1.6.2
networkx: 3.1
nibabel: 5.0.1
nptyping: 2.5.0
numexpr: 2.8.4
numpy: 1.25.1
openvr: 1.23.701
packaging: 21.3
ParmEd: 3.4.3
parso: 0.8.3
pep517: 0.13.0
pexpect: 4.8.0
pickleshare: 0.7.5
Pillow: 10.0.0
pip: 23.0
pkginfo: 1.9.6
platformdirs: 3.9.1
prompt-toolkit: 3.0.39
psutil: 5.9.5
ptyprocess: 0.7.0
pure-eval: 0.2.2
py-cpuinfo: 9.0.0
pycollada: 0.7.2
pydicom: 2.3.0
Pygments: 2.14.0
pynrrd: 1.0.0
PyOpenGL: 3.1.7
PyOpenGL-accelerate: 3.1.7
pyparsing: 3.0.9
pyproject-hooks: 1.0.0
PyQt6: 6.3.1
PyQt6-commercial: 6.3.1
PyQt6-Qt6: 6.3.2
PyQt6-sip: 13.4.0
PyQt6-WebEngine: 6.3.1
PyQt6-WebEngine-commercial: 6.3.1
PyQt6-WebEngine-Qt6: 6.3.2
python-dateutil: 2.8.2
pytz: 2023.3
pyzmq: 25.1.0
qtconsole: 5.4.3
QtPy: 2.3.1
RandomWords: 0.4.0
requests: 2.31.0
scipy: 1.11.1
setuptools: 67.4.0
setuptools-scm: 7.0.5
sfftk-rw: 0.7.3
six: 1.16.0
snowballstemmer: 2.2.0
sortedcontainers: 2.4.0
soupsieve: 2.4.1
sphinx: 6.1.3
sphinx-autodoc-typehints: 1.22
sphinxcontrib-applehelp: 1.0.4
sphinxcontrib-blockdiag: 3.0.0
sphinxcontrib-devhelp: 1.0.2
sphinxcontrib-htmlhelp: 2.0.1
sphinxcontrib-jsmath: 1.0.1
sphinxcontrib-qthelp: 1.0.3
sphinxcontrib-serializinghtml: 1.1.5
stack-data: 0.6.2
tables: 3.8.0
tcia-utils: 1.5.1
tifffile: 2023.7.18
tinyarray: 1.2.4
tomli: 2.0.1
tornado: 6.3.2
traitlets: 5.9.0
typing-extensions: 4.7.1
tzdata: 2023.3
urllib3: 2.0.4
wcwidth: 0.2.6
webcolors: 1.12
wheel: 0.38.4
wheel-filename: 1.4.1
widgetsnbextension: 4.0.8
Attachments (1)
Change History (8)
comment:1 by , 2 years ago
| Component: | Unassigned → Volume Data |
|---|---|
| Owner: | set to |
| Platform: | → all |
| Project: | → ChimeraX |
| Reporter: | changed from to |
| Status: | new → assigned |
| Summary: | ChimeraX bug report submission → Allow fetching fit PDB models together with EMDB maps |
| Type: | defect → enhancement |
comment:2 by , 2 years ago
It turns out that Chimera's ability to fetch fit PDB models with EMDB maps is broken, it won't find any PDB fit models because the EMDB XML metadata changed how it annotates fit models. (Chimera ticket #19739, https://www.rbvi.ucsf.edu/trac/chimera/ticket/19739).
Here is how the current EMDB XML meta identifies fit models from EMDB 34838
https://files.wwpdb.org/pub/emdb/structures/EMD-34838/header/emd-34838.xml
<crossreferences> ... <pdb_list> <pdb_reference> <pdb_id>8hju</pdb_id> <relationship> <in_frame>FULLOVERLAP</in_frame> </relationship> </pdb_reference> </pdb_list> </crossreferences>
comment:3 by , 2 years ago
I will try to allow the open command to fetch both map and fit pdbs with a command like "open 34838 from emdb fits true" or maybe "open emfits:34838". Also I think I will have the Log report the fit PDB identifiers as links when you just open the map, and clicking on the links will open the PDB. These improvements will requires fetching the XML metadata, a small file. That also contains a recommended contour level and I might have ChimeraX use that contour level instead of its current heuristic.
comment:4 by , 2 years ago
| Cc: | added |
|---|---|
| Resolution: | → fixed |
| Status: | assigned → closed |
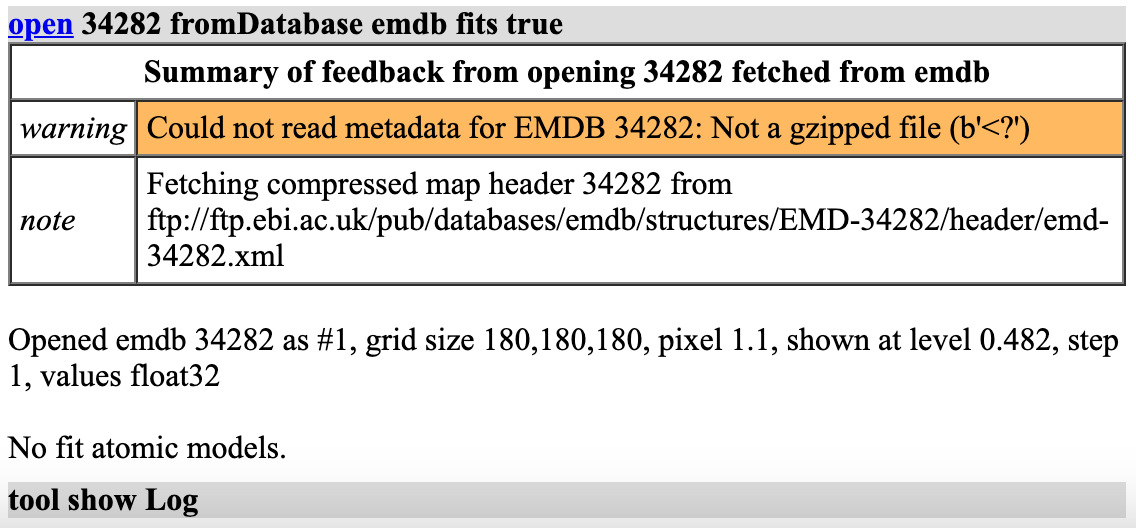
Done.
You can fetch the fit atomic models with EMDB maps using the new "fits" option
open 34282 from emdb fits true
or using the database name emdb_fits
open 34282 from emdb_fits
or using this database name as a prefix
open emdb_fits:34282
All three of these open both the map and the fit atomic models if there are any. Even when you don't open the fits, opening the EMDB map now includes the fit PDB ids as links in the log message and clicking those links will open the PDB (mmCIF format). For example,
open 34282 from emdb
Opened emdb 34282 as #1, grid size 180,180,180, pixel 1.1, shown at level 0.482, step 1, values float32, fit PDB 8gv3
comment:5 by , 2 years ago
This sounds great, thanks Tom!! Oli >> Opened emdb 34282 as #1, grid size 180,180,180, pixel 1.1, shown at
comment:6 by , 2 years ago
comment:7 by , 2 years ago
Fixed.
Not sure how my original code worked since it was asking to uncompress the xml data which was not compressed. Maybe my office iMac had out of date fetching code that allowed it to work.
Also I see that the code used database name emdb_fit instead of what I described as emdb_fits. I've changed it to be emdb_fits.
From: Oliver Clarke via ChimeraX-users <chimerax-users@…>
Subject: [chimerax-users] Re: [Chimera-users] Chimera X menu "View" vs. "Focus"
Date: September 25, 2023 at 10:16:48 AM PDT
To: ChimeraX Users Help <chimerax-users@…>
Cosigned re fetch by id being missed - particularly for EMDB (maps & map+model), which I don’t see an obvious way to access currently even on the command line (apologies if I missed it and it is obvious!)
Cheers
Oli