#7103 closed defect (fixed)
Volume Viewer after session restore: wrapped C/C++ object of type QScrollArea has been deleted
| Reported by: | Owned by: | Tom Goddard | |
|---|---|---|---|
| Priority: | normal | Milestone: | |
| Component: | Sessions | Version: | |
| Keywords: | Cc: | Greg Couch | |
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
The following bug report has been submitted:
Platform: Windows-10-10.0.22000
ChimeraX Version: 1.4 (2022-06-03 23:39:42 UTC)
Description
Opening up any saved ChimeraX session causes this error dialog to occur. Session seems normal and unaffected. Happens with both sessions saved from ChimeraX 1.3 and new sessions created in ChimeraX 1.4.
Also not sure if this makes a difference, my OS is Windows 11 Home (OS build 22000.739), not what the report bug dialog detected in the box below.
Log:
UCSF ChimeraX version: 1.4 (2022-06-03)
© 2016-2022 Regents of the University of California. All rights reserved.
> open
> D:\\\Zhou_Lab\\\Projects\\\HCMV_Tegument\\\Figures\\\Figure_3\\\UL77_interactions_cleft.cxs
Log from Wed Jun 15 14:27:37 2022UCSF ChimeraX version: 1.4 (2022-06-03)
© 2016-2022 Regents of the University of California. All rights reserved.
> open
> D:\\\Zhou_Lab\\\Projects\\\HCMV_Tegument\\\Figures\\\Figure_3\\\UL77_interactions_cleft.cxs
Log from Wed Jun 8 13:59:03 2022UCSF ChimeraX version: 1.3 (2021-12-08)
© 2016-2021 Regents of the University of California. All rights reserved.
> open
> D:\\\Zhou_Lab\\\Projects\\\HCMV_Tegument\\\Figures\\\Figure_3\\\UL77_interactions_cleft.cxs
Log from Wed Jun 8 13:51:01 2022UCSF ChimeraX version: 1.3 (2021-12-08)
© 2016-2021 Regents of the University of California. All rights reserved.
> open
> D:\\\Zhou_Lab\\\Projects\\\HCMV_Tegument\\\Figures\\\Figure_3\\\UL77_interactions_1.cxs
Log from Wed Jun 8 13:06:18 2022UCSF ChimeraX version: 1.3 (2021-12-08)
© 2016-2021 Regents of the University of California. All rights reserved.
> open
> D:\\\Zhou_Lab\\\Projects\\\HCMV_Tegument\\\Figures\\\Figure_3\\\UL77_interactions_1.cxs
Log from Tue Jun 7 03:36:45 2022UCSF ChimeraX version: 1.3 (2021-12-08)
© 2016-2021 Regents of the University of California. All rights reserved.
> open
> D:\\\Zhou_Lab\\\Projects\\\HCMV_Tegument\\\Figures\\\Figure_3\\\UL77_interactions_1.cxs
Log from Fri Jan 14 14:57:10 2022UCSF ChimeraX version: 1.2.5 (2021-05-24)
© 2016-2021 Regents of the University of California. All rights reserved.
How to cite UCSF ChimeraX
> open
> I:/Zhou_Lab/Projects/HCMV_Portal/Modeling/Virion_Type1/UL77_lower_FINAL_VT1_1.pdb
> I:/Zhou_Lab/Projects/HCMV_Portal/Modeling/Virion_Type1/UL77_lower_FINAL_VT1_2.pdb
> I:/Zhou_Lab/Projects/HCMV_Portal/Modeling/Virion_Type1/UL77_lower_FINAL_VT1_3.pdb
> I:/Zhou_Lab/Projects/HCMV_Portal/Modeling/Virion_Type1/UL77_lower_FINAL_VT1_4.pdb
> I:/Zhou_Lab/Projects/HCMV_Portal/Modeling/Virion_Type1/UL77_lower_FINAL_VT1_5.pdb
> I:/Zhou_Lab/Projects/HCMV_Portal/Modeling/Virion_Type1/UL77_lower_pent_combined.pdb
> I:/Zhou_Lab/Projects/HCMV_Portal/Modeling/Virion_Type1/UL77_upper_FINAL_VT1_1.pdb
> I:/Zhou_Lab/Projects/HCMV_Portal/Modeling/Virion_Type1/UL77_upper_FINAL_VT1_2.pdb
> I:/Zhou_Lab/Projects/HCMV_Portal/Modeling/Virion_Type1/UL77_upper_FINAL_VT1_3.pdb
> I:/Zhou_Lab/Projects/HCMV_Portal/Modeling/Virion_Type1/UL77_upper_FINAL_VT1_4.pdb
> I:/Zhou_Lab/Projects/HCMV_Portal/Modeling/Virion_Type1/UL77_upper_FINAL_VT1_5.pdb
> I:/Zhou_Lab/Projects/HCMV_Portal/Modeling/Virion_Type1/UL77_upper_pent_combined.pdb
Summary of feedback from opening
I:/Zhou_Lab/Projects/HCMV_Portal/Modeling/Virion_Type1/UL77_lower_FINAL_VT1_1.pdb
---
warning | Ignored bad PDB record found on line 4038
END
Chain information for UL77_lower_FINAL_VT1_1.pdb #1
---
Chain | Description
A | No description available
Chain information for UL77_lower_FINAL_VT1_2.pdb #2
---
Chain | Description
A | No description available
Chain information for UL77_lower_FINAL_VT1_3.pdb #3
---
Chain | Description
A | No description available
Chain information for UL77_lower_FINAL_VT1_4.pdb #4
---
Chain | Description
A | No description available
Chain information for UL77_lower_FINAL_VT1_5.pdb #5
---
Chain | Description
A | No description available
Chain information for UL77_lower_pent_combined.pdb #6
---
Chain | Description
A B C D E | No description available
Chain information for UL77_upper_FINAL_VT1_1.pdb #7
---
Chain | Description
A | No description available
Chain information for UL77_upper_FINAL_VT1_2.pdb #8
---
Chain | Description
A | No description available
Chain information for UL77_upper_FINAL_VT1_3.pdb #9
---
Chain | Description
A | No description available
Chain information for UL77_upper_FINAL_VT1_4.pdb #10
---
Chain | Description
A | No description available
Chain information for UL77_upper_FINAL_VT1_5.pdb #11
---
Chain | Description
A | No description available
Chain information for UL77_upper_pent_combined.pdb #12
---
Chain | Description
A B C D E | No description available
Drag select of 4925 residues, 90 pseudobonds, 39775 atoms
> hide sel atoms
> show sel cartoons
> select clear
> lighting soft
> lighting simple
> lighting soft
> lighting full
> lighting soft
> lighting simple
> hide #!1 models
> show #!1 models
> dssp
> hide #!1 models
> show #!1 models
> hide #!2-12 target m
> hide #!1 models
> show #!1 models
> show #!2 models
> show #!1-12 target m
> hide #!12 models
> hide #!6 models
> lighting soft
> lighting simple
> lighting soft
> lighting simple
> lighting soft
> set bgColor white
> select clear
> select #11/A:313
7 atoms, 7 bonds, 1 residue, 1 model selected
> select up
138 atoms, 139 bonds, 18 residues, 1 model selected
> select up
3979 atoms, 4082 bonds, 493 residues, 1 model selected
> select clear
> select #11/A:317
8 atoms, 7 bonds, 1 residue, 1 model selected
> select up
138 atoms, 139 bonds, 18 residues, 1 model selected
> select up
3979 atoms, 4082 bonds, 493 residues, 1 model selected
> color sel light sea green
> select #7/A:258
7 atoms, 6 bonds, 1 residue, 1 model selected
> select up
112 atoms, 111 bonds, 15 residues, 1 model selected
> select up
3979 atoms, 4082 bonds, 493 residues, 1 model selected
> color sel forest green
> select clear
> select clear
> select #1/A:255
8 atoms, 7 bonds, 1 residue, 1 model selected
> select up
112 atoms, 111 bonds, 15 residues, 1 model selected
> select up
3976 atoms, 4081 bonds, 492 residues, 1 model selected
> color sel #000055ff
> color sel #0055ffff
> select #10/A:616
6 atoms, 5 bonds, 1 residue, 1 model selected
> select add #9/A:532
15 atoms, 13 bonds, 2 residues, 2 models selected
> select add #8/A:618
23 atoms, 20 bonds, 3 residues, 3 models selected
> select clear
> hide #!2-5 target m
> hide #!8-10 target m
> select #7/A:202
9 atoms, 8 bonds, 1 residue, 1 model selected
> select add #7/A:210
17 atoms, 15 bonds, 2 residues, 1 model selected
> select up
100 atoms, 101 bonds, 12 residues, 1 model selected
> ui tool show "Side View"
> select add #7/A:211
105 atoms, 105 bonds, 13 residues, 1 model selected
> select #7/A:206
7 atoms, 6 bonds, 1 residue, 1 model selected
> select clear
> select add #7/A:210
8 atoms, 7 bonds, 1 residue, 1 model selected
> select add #7/A:208
19 atoms, 18 bonds, 2 residues, 1 model selected
> select clear
> select #7/A:206
7 atoms, 6 bonds, 1 residue, 1 model selected
> select add #7/A:207
15 atoms, 13 bonds, 2 residues, 1 model selected
> select add #7/A:208
26 atoms, 24 bonds, 3 residues, 1 model selected
> select add #7/A:210
34 atoms, 31 bonds, 4 residues, 1 model selected
> select add #7/A:209
43 atoms, 39 bonds, 5 residues, 1 model selected
> select add #7/A:211
48 atoms, 43 bonds, 6 residues, 1 model selected
> select add #7/A:212
54 atoms, 48 bonds, 7 residues, 1 model selected
> select add #7/A:213
61 atoms, 54 bonds, 8 residues, 1 model selected
> show sel atoms
> style sel stick
Changed 61 atom styles
> color sel byhetero
> select #11/A:563
6 atoms, 5 bonds, 1 residue, 1 model selected
> select add #11/A:564
14 atoms, 12 bonds, 2 residues, 1 model selected
> select add #11/A:532
23 atoms, 20 bonds, 3 residues, 1 model selected
> select add #11/A:613
30 atoms, 26 bonds, 4 residues, 1 model selected
> select add #11/A:224
41 atoms, 36 bonds, 5 residues, 1 model selected
> select up
416 atoms, 423 bonds, 51 residues, 1 model selected
> select subtract #11/A:525
408 atoms, 416 bonds, 50 residues, 1 model selected
> select subtract #11/A:524
404 atoms, 413 bonds, 49 residues, 1 model selected
> select subtract #11/A:526
399 atoms, 409 bonds, 48 residues, 1 model selected
> select subtract #11/A:527
391 atoms, 402 bonds, 47 residues, 1 model selected
> show sel atoms
> hide sel atoms
> select subtract #11/A:536
385 atoms, 397 bonds, 46 residues, 1 model selected
> select subtract #11/A:537
381 atoms, 394 bonds, 45 residues, 1 model selected
> select add #11/A:536
387 atoms, 399 bonds, 46 residues, 1 model selected
> select subtract #11/A:538
373 atoms, 384 bonds, 45 residues, 1 model selected
> select subtract #11/A:539
365 atoms, 377 bonds, 44 residues, 1 model selected
> select subtract #11/A:540
358 atoms, 370 bonds, 43 residues, 1 model selected
> select subtract #11/A:541
352 atoms, 365 bonds, 42 residues, 1 model selected
> select subtract #11/A:542
341 atoms, 355 bonds, 41 residues, 1 model selected
> select subtract #11/A:543
330 atoms, 345 bonds, 40 residues, 1 model selected
> select subtract #11/A:544
321 atoms, 337 bonds, 39 residues, 1 model selected
> select subtract #11/A:545
313 atoms, 330 bonds, 38 residues, 1 model selected
> show sel atoms
> hide sel atoms
> select subtract #11/A:607
306 atoms, 323 bonds, 37 residues, 1 model selected
> select subtract #11/A:609
296 atoms, 313 bonds, 36 residues, 1 model selected
> select subtract #11/A:610
285 atoms, 303 bonds, 35 residues, 1 model selected
> select subtract #11/A:608
274 atoms, 293 bonds, 34 residues, 1 model selected
> select subtract #11/A:611
267 atoms, 287 bonds, 33 residues, 1 model selected
> select subtract #11/A:612
261 atoms, 282 bonds, 32 residues, 1 model selected
> show sel atoms
> color sel byhetero
> hide sel atoms
> select subtract #11/A:536
255 atoms, 277 bonds, 31 residues, 1 model selected
> show sel atoms
> select subtract #11/A:533
249 atoms, 272 bonds, 30 residues, 1 model selected
> select subtract #11/A:532
240 atoms, 264 bonds, 29 residues, 1 model selected
> select subtract #11/A:529
235 atoms, 260 bonds, 28 residues, 1 model selected
> select subtract #11/A:528
227 atoms, 253 bonds, 27 residues, 1 model selected
> hide sel atoms
> show sel atoms
> select add #11/A:533
233 atoms, 258 bonds, 28 residues, 1 model selected
> select add #11/A:529
238 atoms, 262 bonds, 29 residues, 1 model selected
> select add #11/A:528
246 atoms, 269 bonds, 30 residues, 1 model selected
> hide sel atoms
> select add #11/A:532
255 atoms, 277 bonds, 31 residues, 1 model selected
> hide sel atoms
> select subtract #11/A:532
246 atoms, 269 bonds, 30 residues, 1 model selected
> select subtract #11/A:533
240 atoms, 264 bonds, 29 residues, 1 model selected
> select subtract #11/A:529
235 atoms, 260 bonds, 28 residues, 1 model selected
> show sel atoms
> hide sel atoms
> select subtract #11/A:528
227 atoms, 253 bonds, 27 residues, 1 model selected
> show sel atoms
> hide sel atoms
> show sel atoms
> hide sel atoms
> select subtract #11/A:619
216 atoms, 242 bonds, 26 residues, 1 model selected
> select subtract #11/A:222
204 atoms, 230 bonds, 25 residues, 1 model selected
> show sel atoms
> select clear
Drag select of 81 residues
> hide sel target a
> hide sel cartoons
> show sel cartoons
> select clear
> select #11/A:613
7 atoms, 6 bonds, 1 residue, 1 model selected
> select add #7/A:251
16 atoms, 14 bonds, 2 residues, 2 models selected
> select up
214 atoms, 215 bonds, 28 residues, 2 models selected
> select up
7958 atoms, 8164 bonds, 986 residues, 2 models selected
> style sel sphere
Changed 7958 atom styles
> style sel ball
Changed 7958 atom styles
> style sel stick
Changed 7958 atom styles
> color sel bychain
> undo
> hide sel atoms
> show sel atoms
> hide sel atoms
> show sel atoms
> hide sel atoms
> undo
> undo
> undo
> undo
> select clear
> select #7/A:208
11 atoms, 11 bonds, 1 residue, 1 model selected
> select add #7/A:209
20 atoms, 19 bonds, 2 residues, 1 model selected
> select add #7/A:210
28 atoms, 26 bonds, 3 residues, 1 model selected
> select add #7/A:211
33 atoms, 30 bonds, 4 residues, 1 model selected
> select add #7/A:212
39 atoms, 35 bonds, 5 residues, 1 model selected
> select add #7/A:213
46 atoms, 41 bonds, 6 residues, 1 model selected
> select add #7/A:214
60 atoms, 56 bonds, 7 residues, 1 model selected
> show sel atoms
> select #7/A:206
7 atoms, 6 bonds, 1 residue, 1 model selected
> show sel cartoons
> show sel atoms
> select #7/A:208
11 atoms, 11 bonds, 1 residue, 1 model selected
> hide sel atoms
> show sel atoms
> select clear
Drag select of 1 residues
> select clear
> select #11/A:227
11 atoms, 10 bonds, 1 residue, 1 model selected
> select add #11/A:224
22 atoms, 20 bonds, 2 residues, 1 model selected
> select add #11/A:220
30 atoms, 27 bonds, 3 residues, 1 model selected
> select add #11/A:221
44 atoms, 42 bonds, 4 residues, 1 model selected
> select add #11/A:228
48 atoms, 45 bonds, 5 residues, 1 model selected
> select add #11/A:225
56 atoms, 52 bonds, 6 residues, 1 model selected
> select add #11/A:223
68 atoms, 64 bonds, 7 residues, 1 model selected
> show sel atoms
> hide #!1 models
> select #7/A:205
8 atoms, 7 bonds, 1 residue, 1 model selected
> show sel atoms
> hide sel atoms
> select #7/A:215
7 atoms, 6 bonds, 1 residue, 1 model selected
> show sel atoms
> color sel byhetero
> select clear
> select #7/A:214
14 atoms, 15 bonds, 1 residue, 1 model selected
> color sel byhetero
> select clear
> select #7/A:207
8 atoms, 7 bonds, 1 residue, 1 model selected
> select #11/A:617
6 atoms, 5 bonds, 1 residue, 1 model selected
> select up
102 atoms, 104 bonds, 13 residues, 1 model selected
> select up
3979 atoms, 4082 bonds, 493 residues, 1 model selected
> show sel atoms
> select add #7/A:464
3987 atoms, 4089 bonds, 494 residues, 2 models selected
> select up
4020 atoms, 4122 bonds, 499 residues, 2 models selected
> select up
7958 atoms, 8164 bonds, 986 residues, 2 models selected
> select clear
> select #7/A:464
8 atoms, 7 bonds, 1 residue, 1 model selected
> select add #11/A:616
14 atoms, 12 bonds, 2 residues, 2 models selected
> select up
143 atoms, 144 bonds, 19 residues, 2 models selected
> select up
7958 atoms, 8164 bonds, 986 residues, 2 models selected
> show sel atoms
> color sel byhetero
> hide sel atoms
> select clear
> select #7/A:206
7 atoms, 6 bonds, 1 residue, 1 model selected
> select add #7/A:210
15 atoms, 13 bonds, 2 residues, 1 model selected
> select add #7/A:209
24 atoms, 21 bonds, 3 residues, 1 model selected
> select add #7/A:211
29 atoms, 25 bonds, 4 residues, 1 model selected
> select add #7/A:212
35 atoms, 30 bonds, 5 residues, 1 model selected
> select add #7/A:213
42 atoms, 36 bonds, 6 residues, 1 model selected
> select add #7/A:214
56 atoms, 51 bonds, 7 residues, 1 model selected
> select add #7/A:215
63 atoms, 57 bonds, 8 residues, 1 model selected
> select add #11/A:617
69 atoms, 62 bonds, 9 residues, 2 models selected
> select add #11/A:220
77 atoms, 69 bonds, 10 residues, 2 models selected
> select add #11/A:221
91 atoms, 84 bonds, 11 residues, 2 models selected
> select add #11/A:224
102 atoms, 94 bonds, 12 residues, 2 models selected
> select add #11/A:223
114 atoms, 106 bonds, 13 residues, 2 models selected
> select subtract #11/A:223
102 atoms, 94 bonds, 12 residues, 2 models selected
> select add #11/A:225
110 atoms, 101 bonds, 13 residues, 2 models selected
> select add #11/A:227
121 atoms, 111 bonds, 14 residues, 2 models selected
> select add #11/A:228
125 atoms, 114 bonds, 15 residues, 2 models selected
> select add #11/A:231
132 atoms, 120 bonds, 16 residues, 2 models selected
> show sel atoms
> select clear
> select #11/A:564
8 atoms, 7 bonds, 1 residue, 1 model selected
> select add #11/A:563
14 atoms, 12 bonds, 2 residues, 1 model selected
> select add #11/A:562
23 atoms, 20 bonds, 3 residues, 1 model selected
> show sel atoms
> select #7/A:215
7 atoms, 6 bonds, 1 residue, 1 model selected
> hide sel atoms
> select #11/A:245
8 atoms, 7 bonds, 1 residue, 1 model selected
> select add #11/A:246
16 atoms, 14 bonds, 2 residues, 1 model selected
> select add #11/A:247
27 atoms, 25 bonds, 3 residues, 1 model selected
> show sel atoms
> select #11/A:223
12 atoms, 12 bonds, 1 residue, 1 model selected
> show sel atoms
> select #11/A:247
11 atoms, 11 bonds, 1 residue, 1 model selected
> select add #11/A:246
19 atoms, 18 bonds, 2 residues, 1 model selected
> select add #11/A:245
27 atoms, 25 bonds, 3 residues, 1 model selected
> hide sel atoms
> select clear
> select #11/A:230
12 atoms, 12 bonds, 1 residue, 1 model selected
> show sel atoms
> hide sel atoms
> select #7/A:208
11 atoms, 11 bonds, 1 residue, 1 model selected
> show sel atoms
> hide sel atoms
> show sel atoms
> select #11/A:617
6 atoms, 5 bonds, 1 residue, 1 model selected
> hide sel cartoons
> show sel cartoons
> hide sel atoms
> select add #11/A:614
14 atoms, 12 bonds, 2 residues, 1 model selected
> select add #11/A:613
21 atoms, 18 bonds, 3 residues, 1 model selected
> show sel atoms
> hide sel atoms
> select clear
> select #11/A:535
11 atoms, 10 bonds, 1 residue, 1 model selected
> select add #7/A:470
16 atoms, 14 bonds, 2 residues, 2 models selected
> select add #11/A:536
22 atoms, 19 bonds, 3 residues, 2 models selected
> select add #7/A:469
30 atoms, 26 bonds, 4 residues, 2 models selected
> show sel atoms
> hide sel atoms
> show sel atoms
> select #7/A:469
8 atoms, 7 bonds, 1 residue, 1 model selected
> select clear
> select #7/A:469
8 atoms, 7 bonds, 1 residue, 1 model selected
> hide sel atoms
> select #11/A:536
6 atoms, 5 bonds, 1 residue, 1 model selected
> hide sel atoms
> view name close1
> view list
Named views: close1
> view close1
> select #11/A:563
6 atoms, 5 bonds, 1 residue, 1 model selected
> select #11/A:562
9 atoms, 8 bonds, 1 residue, 1 model selected
> hide sel atoms
> select #7/A:467
4 atoms, 3 bonds, 1 residue, 1 model selected
> select add #7/A:466
12 atoms, 10 bonds, 2 residues, 1 model selected
> show sel atoms
> select #7/A:470
5 atoms, 4 bonds, 1 residue, 1 model selected
> hide sel atoms
> select #7/A:469
8 atoms, 7 bonds, 1 residue, 1 model selected
> show sel atoms
> view list
Named views: close1
> view close1
> hide sel atoms
> select clear
> select #7/A:466
8 atoms, 7 bonds, 1 residue, 1 model selected
> hide sel atoms
> select #7/A:206
7 atoms, 6 bonds, 1 residue, 1 model selected
> hide sel atoms
> select #7/A:211
5 atoms, 4 bonds, 1 residue, 1 model selected
> hide sel atoms
> show sel atoms
> view close1
> view close1
> view close1
> select clear
> select #7/A:206
7 atoms, 6 bonds, 1 residue, 1 model selected
> select add #7/A:205
15 atoms, 13 bonds, 2 residues, 1 model selected
> show sel atoms
> select #7/A:202
9 atoms, 8 bonds, 1 residue, 1 model selected
> select add #11/A:561
15 atoms, 13 bonds, 2 residues, 2 models selected
> show sel cartoons
> show sel atoms
> select clear
> select #11/A:561
6 atoms, 5 bonds, 1 residue, 1 model selected
> hide sel atoms
> select #11/A:562
9 atoms, 8 bonds, 1 residue, 1 model selected
> show sel atoms
> view list
Named views: close1
> view close1
> view close1
> view close1
> save
> I:/Zhou_Lab/Projects/HCMV_Portal/Figures/Figure_3/UL77_interactions_1.cxs
——— End of log from Fri Jan 14 14:57:10 2022 ———
opened ChimeraX session
> ui tool show "Side View"
> select clear
> open
> D:/Zhou_Lab/Projects/HCMV_Tegument/Modeling/Virion_Type1/AsymTEG_VT1_1.pdb
Summary of feedback from opening
D:/Zhou_Lab/Projects/HCMV_Tegument/Modeling/Virion_Type1/AsymTEG_VT1_1.pdb
---
warnings | Ignored bad PDB record found on line 541
METHOD ROSETTA provides both ab initio and
Ignored bad PDB record found on line 542
METHOD comparative models of protein domains.
Ignored bad PDB record found on line 543
METHOD Comparative models are built from structures
Ignored bad PDB record found on line 544
METHOD detected and aligned by HHSEARCH, SPARKS, and
Ignored bad PDB record found on line 545
METHOD Raptor. Loop regions are assembled from fragments and
5 messages similar to the above omitted
Chain information for AsymTEG_VT1_1.pdb #13
---
Chain | Description
A | No description available
B | No description available
C | No description available
D | No description available
E | No description available
F | No description available
G | No description available
> open
> D:/Zhou_Lab/Projects/HCMV_Tegument/Modeling/Virion_Type1/AsymTEG_VT1_5.pdb
Summary of feedback from opening
D:/Zhou_Lab/Projects/HCMV_Tegument/Modeling/Virion_Type1/AsymTEG_VT1_5.pdb
---
warnings | Ignored bad PDB record found on line 541
METHOD ROSETTA provides both ab initio and
Ignored bad PDB record found on line 542
METHOD comparative models of protein domains.
Ignored bad PDB record found on line 543
METHOD Comparative models are built from structures
Ignored bad PDB record found on line 544
METHOD detected and aligned by HHSEARCH, SPARKS, and
Ignored bad PDB record found on line 545
METHOD Raptor. Loop regions are assembled from fragments and
5 messages similar to the above omitted
Chain information for AsymTEG_VT1_5.pdb #14
---
Chain | Description
A | No description available
B | No description available
C | No description available
D | No description available
E | No description available
F | No description available
G | No description available
> hide #!7,11,13-14 atoms
> show #!7,11,13-14 cartoons
> hide #!7 models
> hide #!11 models
> select clear
Drag select of 86 residues
> select add #13/F:567
724 atoms, 4 bonds, 87 residues, 2 models selected
> select up
1411 atoms, 1448 bonds, 175 residues, 2 models selected
> select up
11778 atoms, 12083 bonds, 1451 residues, 2 models selected
> select ~sel
130270 atoms, 133549 bonds, 123 pseudobonds, 16107 residues, 28 models
selected
> delete atoms (#!13-14 & sel)
> delete bonds (#!13-14 & sel)
> select clear
> color #13/E crimson
> select clear
> select #13/F:236
9 atoms, 8 bonds, 1 residue, 1 model selected
> select up
71 atoms, 74 bonds, 7 residues, 1 model selected
> select up
4000 atoms, 4105 bonds, 495 residues, 1 model selected
> hide sel cartoons
> show sel cartoons
> select clear
> view list
Named views: close1
> view close1
[Repeated 3 time(s)]
> select #14/E:224
11 atoms, 10 bonds, 1 residue, 1 model selected
> select add #14/E:617
17 atoms, 15 bonds, 2 residues, 1 model selected
> select add #13/E:209
26 atoms, 23 bonds, 3 residues, 2 models selected
> select up
318 atoms, 325 bonds, 37 residues, 2 models selected
> select add #13/E:213
325 atoms, 331 bonds, 38 residues, 2 models selected
> select up
389 atoms, 400 bonds, 46 residues, 2 models selected
> show sel atoms
> style sel stick
Changed 389 atom styles
> hide #!13 models
> hide #!14 models
> select clear
> show #!7 models
> show #!11 models
> select #7/A:212
6 atoms, 5 bonds, 1 residue, 1 model selected
> select add #11/A:223
18 atoms, 17 bonds, 2 residues, 2 models selected
> select up
187 atoms, 194 bonds, 21 residues, 2 models selected
> show sel atoms
> show #!13 models
> hide #!13 models
> show #!14 models
> hide #!14 models
> select clear
> hide #!7 models
> hide #!11 models
> show #!14 models
> show #!13 models
> select #14/E:564
8 atoms, 7 bonds, 1 residue, 1 model selected
> select up
79 atoms, 80 bonds, 10 residues, 1 model selected
> show sel atoms
> style sel stick
Changed 79 atom styles
> view list
Named views: close1
> view close1
> hide sel atoms
> select clear
> hide #!13-14 atoms
> view close1
> select clear
> select #13-14/E-F:1-150
2125 atoms, 2154 bonds, 1 pseudobond, 256 residues, 3 models selected
> hide sel atoms
> hide sel cartoons
> select clear
> select #13-14/E-F:1-150
2125 atoms, 2154 bonds, 1 pseudobond, 256 residues, 3 models selected
> show sel cartoons
> cofr frontCenter
[Repeated 1 time(s)]
> select clear
[Repeated 1 time(s)]
> select #13/F:212
6 atoms, 5 bonds, 1 residue, 1 model selected
> select clear
> select #13/F:212
6 atoms, 5 bonds, 1 residue, 1 model selected
> select add #14/E:360
10 atoms, 8 bonds, 2 residues, 2 models selected
> select add #14/E:342
16 atoms, 13 bonds, 3 residues, 2 models selected
> select add #14/E:384
20 atoms, 16 bonds, 4 residues, 2 models selected
> select add #13/F:209
29 atoms, 24 bonds, 5 residues, 2 models selected
> select up
536 atoms, 554 bonds, 64 residues, 2 models selected
> show sel atoms
> style sel stick
Changed 536 atom styles
> hide sel atoms
> select clear
> select #13-14/E-F:1-150
2125 atoms, 2154 bonds, 1 pseudobond, 256 residues, 3 models selected
> hide sel cartoons
> select clear
> cofr frontCenter
> view name far1
> view list
Named views: close1, far1
> view close1
> view far1
[Repeated 12 time(s)]
> view name far2
> view list
Named views: close1, far1, far2
> view far1
> view far2
> view far1
> view far2
> select #13-14/E-F:1-150
2125 atoms, 2154 bonds, 1 pseudobond, 256 residues, 3 models selected
> show sel cartoons
> hide sel cartoons
> show sel cartoons
> set bgColor black
> set bgColor white
> view list
Named views: close1, far1, far2
> view far2
> view far1
> view far2
> view far1
> view far2
> view far1
> view far2
> select clear
> view far2
> color #13/F pink
> color #14/E steel blue
> color #14/E #d5e68d
> color #14/E #8fe2c1
> color #13/F #d5e68d
> color #13/F pink
> color #13/F #d5e68d
> select #13/F:612
6 atoms, 5 bonds, 1 residue, 1 model selected
> ui tool show "Selection Inspector"
> color #13/E #d5e68d
> select #13/E:528
8 atoms, 7 bonds, 1 residue, 1 model selected
> ui tool show "Selection Inspector"
> color sel #737c4c atoms
> color #13/E #737c4c
> select clear
> color #13/F pink
> color #13/E crimson
> color #13/F pink
> color #14/E #558672
> color #14/E slate blue
> color #14/E #8fe2c1
> color #14/E turquoise
> view list
Named views: close1, far1, far2
> view far2
> view far1
> view far2
> view far1
> view far2
> color #14/E #8fe2c1
> view close1
> view far2
> color #14/E slate blue
> view close1
> color #13/E:208-210,251-252 gold
> color #13/E:208-210,251-252,464-469 gold
> select #14/E:531
7 atoms, 6 bonds, 1 residue, 1 model selected
> select add #14/E:535
18 atoms, 16 bonds, 2 residues, 1 model selected
> select add #14/E:609
28 atoms, 26 bonds, 3 residues, 1 model selected
> select add #14/E:617
34 atoms, 31 bonds, 4 residues, 1 model selected
> select add #14/E:613
41 atoms, 37 bonds, 5 residues, 1 model selected
> select add #14/E:220
49 atoms, 44 bonds, 6 residues, 1 model selected
> select add #14/E:224
60 atoms, 54 bonds, 7 residues, 1 model selected
> select add #14/E:227
71 atoms, 64 bonds, 8 residues, 1 model selected
> select add #14/E:228
75 atoms, 67 bonds, 9 residues, 1 model selected
> color sel #8fe2c1
> select clear
> select #14/E:609
10 atoms, 10 bonds, 1 residue, 1 model selected
> select add #14/E:220
18 atoms, 17 bonds, 2 residues, 1 model selected
> select add #14/E:535
29 atoms, 27 bonds, 3 residues, 1 model selected
> select up
310 atoms, 316 bonds, 38 residues, 1 model selected
> color sel #8fe2c1
> select clear
> undo
> show sel atoms
> style sel stick
Changed 310 atom styles
> color #13/E:208-210,251-252,464-469 gold
> select #13/E:208-210,251-252,464-469
74 atoms, 72 bonds, 9 residues, 1 model selected
> show sel atoms
> style sel stick
Changed 74 atom styles
> select clear
> hide #!14 models
> hide #!13 models
> show #!11 models
> show #!7 models
> hide #!11 models
> hide #!7 models
> show #!13 models
> show #!14 models
> view list
Named views: close1, far1, far2
> view far2
> color #14/E slate blue
> hide #!13-14 atoms
> color #13/E crimson
> color
> #14/E:610,614,618,221,224,225,227,228,231,232,527,530,531,532,534-536,562-567
> #8fe2c1
> color #13/E:251,252,205,206,209,210,466,469 orange
> color #13/E:251,252,205,206,209,210,466,469 #d5e68d
> view close1
> view far2
> select #13/E:251,252,205,206,209,210,466,469
58 atoms, 54 bonds, 7 residues, 1 model selected
> style sel sphere
Changed 58 atom styles
> show sel atoms
> select
> #14/E:610,614,618,221,224,225,227,228,231,232,527,530,531,532,534-536,562-567
188 atoms, 180 bonds, 23 residues, 1 model selected
> show sel atoms
> style sel sphere
Changed 188 atom styles
> select clear
> view far2
> select
> #14/E:610,614,618,221,224,225,227,228,231,232,527,530,531,532,534-536,562-567
188 atoms, 180 bonds, 23 residues, 1 model selected
> hide sel atoms
> hide sel cartoons
> show sel cartoons
> style sel stick
Changed 188 atom styles
> hide sel cartoons
> style sel stick
Changed 188 atom styles
> show sel atoms
> hide sel atoms
> show sel cartoons
> style sel stick
Changed 188 atom styles
> hide sel cartoons
> style sel stick
Changed 188 atom styles
> show sel cartoons
> show sel atoms
> hide sel cartoons
> hide sel atoms
> show sel atoms
> hide sel atoms
> show sel cartoons
> show sel atoms
> style sel sphere
Changed 188 atom styles
> hide sel atoms
Drag select of 37 atoms, 248 residues, 2 pseudobonds
> hide sel atoms
> select clear
> view list
Named views: close1, far1, far2
> view close1
> view far2
> color #14/E #8fe2c1
> select clear
> color #13/E crimson
> color #14/E slate blue
> save C:\Users\jihma\Desktop\image5.png supersample 3
> hide #!14 models
> show #!14 models
> hide #!13 models
> show #!13 models
> select #13/E:511
5 atoms, 4 bonds, 1 residue, 1 model selected
> select up
53 atoms, 56 bonds, 6 residues, 1 model selected
> select up
3889 atoms, 3989 bonds, 478 residues, 1 model selected
> hide sel cartoons
> save C:\Users\jihma\Desktop\image6.png supersample 3
> show sel cartoons
> select clear
> view list
Named views: close1, far1, far2
> view far2
> select #13/E:181
7 atoms, 7 bonds, 1 residue, 1 model selected
> select up
209 atoms, 214 bonds, 25 residues, 1 model selected
> select up
3889 atoms, 3989 bonds, 478 residues, 1 model selected
> hide sel cartoons
> save C:\Users\jihma\Desktop\image6.png supersample 3
> color #14/E green
> color #14/E forest green
> color #14/E #8fe2c1
> select clear
> save C:\Users\jihma\Desktop\image10.png supersample 3
> view far2
> show #!13-14 cartoons
> select clear
> save C:\Users\jihma\Desktop\image11.png supersample 3
> preset cartoons/nucleotides cylinders/stubs
Changed 50629 atom styles
Preset expands to these ChimeraX commands:
show nucleic
hide protein|solvent|H
surf hide
style (protein|nucleic|solvent) & @@draw_mode=0 stick
cartoon
cartoon style modeh def arrows t arrowshelix f arrowscale 2 wid 2 thick 0.4 sides 12 div 20
cartoon style ~(nucleic|strand) x round
cartoon style (nucleic|strand) x rect
cartoon style protein modeh tube rad 2 sides 24 thick 0.6
cartoon style nucleic x round width 1.6 thick 1.6
nucleotides stubs
> preset cartoons/nucleotides ribbons/slabs
Changed 0 atom styles
Preset expands to these ChimeraX commands:
show nucleic
hide protein|solvent|H
surf hide
style (protein|nucleic|solvent) & @@draw_mode=0 stick
cartoon
cartoon style modeh def arrows t arrowshelix f arrowscale 2 wid 2 thick 0.4 sides 12 div 20
cartoon style ~(nucleic|strand) x round
cartoon style (nucleic|strand) x rect
nucleotides tube/slab shape box
> preset cartoons/nucleotides cylinders/stubs
Changed 0 atom styles
Preset expands to these ChimeraX commands:
show nucleic
hide protein|solvent|H
surf hide
style (protein|nucleic|solvent) & @@draw_mode=0 stick
cartoon
cartoon style modeh def arrows t arrowshelix f arrowscale 2 wid 2 thick 0.4 sides 12 div 20
cartoon style ~(nucleic|strand) x round
cartoon style (nucleic|strand) x rect
cartoon style protein modeh tube rad 2 sides 24 thick 0.6
cartoon style nucleic x round width 1.6 thick 1.6
nucleotides stubs
> preset cartoons/nucleotides licorice/ovals
Changed 0 atom styles
Preset expands to these ChimeraX commands:
show nucleic
hide protein|solvent|H
surf hide
style (protein|nucleic|solvent) & @@draw_mode=0 stick
cartoon
cartoon style modeh def arrows t arrowshelix f arrowscale 2 wid 2 thick 0.4 sides 12 div 20
cartoon style ~(nucleic|strand) x round
cartoon style (nucleic|strand) x rect
cartoon style protein modeh default arrows f x round width 1 thick 1
cartoon style nucleic x round width 1.6 thick 1.6
nucleotides tube/slab shape ellipsoid
> preset cartoons/nucleotides ribbons/slabs
Changed 0 atom styles
Preset expands to these ChimeraX commands:
show nucleic
hide protein|solvent|H
surf hide
style (protein|nucleic|solvent) & @@draw_mode=0 stick
cartoon
cartoon style modeh def arrows t arrowshelix f arrowscale 2 wid 2 thick 0.4 sides 12 div 20
cartoon style ~(nucleic|strand) x round
cartoon style (nucleic|strand) x rect
nucleotides tube/slab shape box
> view list
Named views: close1, far1, far2
> view far1
> view far2
> view close1
> view far2
> save
> D:/Zhou_Lab/Projects/HCMV_Tegument/Figures/Figure_3/UL77_interactions_1.cxs
——— End of log from Tue Jun 7 03:36:45 2022 ———
opened ChimeraX session
> close #13
> open
> D:/Zhou_Lab/Projects/HCMV_Tegument/Modeling/Virion_Type1/AsymTEG_VT1_1.pdb
Summary of feedback from opening
D:/Zhou_Lab/Projects/HCMV_Tegument/Modeling/Virion_Type1/AsymTEG_VT1_1.pdb
---
warnings | Ignored bad PDB record found on line 541
METHOD ROSETTA provides both ab initio and
Ignored bad PDB record found on line 542
METHOD comparative models of protein domains.
Ignored bad PDB record found on line 543
METHOD Comparative models are built from structures
Ignored bad PDB record found on line 544
METHOD detected and aligned by HHSEARCH, SPARKS, and
Ignored bad PDB record found on line 545
METHOD Raptor. Loop regions are assembled from fragments and
5 messages similar to the above omitted
Chain information for AsymTEG_VT1_1.pdb #13
---
Chain | Description
A | No description available
B | No description available
C | No description available
D | No description available
E | No description available
F | No description available
G | No description available
> hide #!14 models
> hide #!13 models
> show #!13 models
> select #13/E,F
7915 atoms, 8120 bonds, 11 pseudobonds, 977 residues, 2 models selected
> select ~sel
106799 atoms, 109526 bonds, 110 pseudobonds, 13209 residues, 28 models
selected
> delete atoms (#!13 & sel)
> delete bonds (#!13 & sel)
> hide #!13 atoms
> show #!13 cartoons
> color #13/E crimson
> color #13/F pink
> show #!14 models
> select clear
[Repeated 1 time(s)]
> preset cartoons/nucleotides cylinders/stubs
Changed 7915 atom styles
Preset expands to these ChimeraX commands:
show nucleic
hide protein|solvent|H
surf hide
style (protein|nucleic|solvent) & @@draw_mode=0 stick
cartoon
cartoon style modeh def arrows t arrowshelix f arrowscale 2 wid 2 thick 0.4 sides 12 div 20
cartoon style ~(nucleic|strand) x round
cartoon style (nucleic|strand) x rect
cartoon style protein modeh tube rad 2 sides 24 thick 0.6
cartoon style nucleic x round width 1.6 thick 1.6
nucleotides stubs
> ui tool show "Side View"
> view list
Named views: close1, far1, far2
> view far1
> view far2
> view far1
> view far2
[Repeated 2 time(s)]
> lighting simple
> lighting full
> lighting soft
> lighting shadows true intensity 0.5
> lighting shadows false
> lighting shadows true
> lighting simple
> lighting soft
> view name far1
> save
> D:/Zhou_Lab/Projects/HCMV_Tegument/Figures/Figure_3/UL77_interactions_1.cxs
——— End of log from Wed Jun 8 13:06:18 2022 ———
opened ChimeraX session
> view list
Named views: close1, far1, far2
> view close1
> preset cartoons/nucleotides ribbons/slabs
Changed 0 atom styles
Preset expands to these ChimeraX commands:
show nucleic
hide protein|solvent|H
surf hide
style (protein|nucleic|solvent) & @@draw_mode=0 stick
cartoon
cartoon style modeh def arrows t arrowshelix f arrowscale 2 wid 2 thick 0.4 sides 12 div 20
cartoon style ~(nucleic|strand) x round
cartoon style (nucleic|strand) x rect
nucleotides tube/slab shape box
> select #13/E:251,252,205,206,209,210,466,469
66 atoms, 61 bonds, 8 residues, 1 model selected
> color sel lawn green
> view close1
> show sel atoms
> style sel ball
Changed 66 atom styles
> hide sel atoms
> select clear
> color #14/E:610,614,618,221,224,225,227,228,530,531,532,534-536,562-567
> select clear
> select #14/E:610,614,618,221,224,225,227,228,530,531,532,534-536,562-567
164 atoms, 158 bonds, 20 residues, 1 model selected
> color sel orange
> select clear
> color #14/E #8fe2c1
> color #13/E crimson
> redo
[Repeated 1 time(s)]
> undo
[Repeated 1 time(s)]
> select #14/E:610,614,618,221,224,225,227,228,530,531,532,534-536,562-567
164 atoms, 158 bonds, 20 residues, 1 model selected
> color #13/E crimson
> hide sel atoms
> select clear
> color #14/E #8fe2c1
> select #14/E:610,614,618,221,224,225,227,228,530,531,532,534-536,562-567
164 atoms, 158 bonds, 20 residues, 1 model selected
> show sel atoms
> style sel ball
Changed 164 atom styles
> color sel byhetero
> view close1
> hide sel atoms
> show sel atoms
> style sel sphere
Changed 164 atom styles
> undo
> select up
467 atoms, 478 bonds, 58 residues, 1 model selected
> select clear
> select #14/E:610,614,618,221,224,225,227,228,530,531,532,534-536,562-567
164 atoms, 158 bonds, 20 residues, 1 model selected
> select #13/E:251,252,205,206,209,210,466,469
66 atoms, 61 bonds, 8 residues, 1 model selected
> select #14/E:610,614,618,221,224,225,227,228,530,531,532,534-536,562-567
164 atoms, 158 bonds, 20 residues, 1 model selected
> select add #13/E:251,252,205,206,209,210,466,469
230 atoms, 219 bonds, 28 residues, 2 models selected
> hide sel atoms
> select clear
> view close1
[Repeated 1 time(s)]
> select #14/E:610,614,618,221,224,225,227,228,530,531,532,534-536,562-567
164 atoms, 158 bonds, 20 residues, 1 model selected
> select add #13/E:251,252,205,206,209,210,466,469
230 atoms, 219 bonds, 28 residues, 2 models selected
> show sel atoms
> style sel stick
Changed 230 atom styles
> style sel sphere
Changed 230 atom styles
> style sel ball
Changed 230 atom styles
> style sel stick
Changed 230 atom styles
> color sel by hetero
Expected a color or one of 'byatom', 'bychain', 'byelement', 'byhetero',
'byidentity', 'bymodel', 'bynucleotide', 'bypolymer', 'fromatoms', or 'random'
or a keyword
> color sel byhetero
> select clear
> undo
> style sel sphere
Changed 230 atom styles
> undo
> style sel ball
Changed 230 atom styles
> select clear
> ui tool show "Side View"
> view list
Named views: close1, far1, far2
> view name close2
> view list
Named views: close1, close2, far1, far2
> view close1
> view close2
> view close1
> view close2
> view name close2
> view list
Named views: close1, close2, far1, far2
> view close2
[Repeated 1 time(s)]
> select #14/E:610,614,618,221,224,225,227,228,530,531,532,534-536,562-567
164 atoms, 158 bonds, 20 residues, 1 model selected
> select add #13/E:251,252,205,206,209,210,466,469
230 atoms, 219 bonds, 28 residues, 2 models selected
> color #14/E #8fe2c1
> color #13/E crimson
> select #14/E:610,614,618,221,224,225,227,228,530,531,532,534-536,562-567
164 atoms, 158 bonds, 20 residues, 1 model selected
> select add #13/E:251,252,205,206,209,210,466,469
230 atoms, 219 bonds, 28 residues, 2 models selected
> style sel sphere
Changed 230 atom styles
> transparency 50
[Repeated 1 time(s)]
> style sel ball
Changed 230 atom styles
> color #14/E:610,614,618,221,224,225,227,228,530,531,532,534-536,562-567
> orange
> color #13/E:251,252,205,206,209,210,466,469 lawn green
> hide sel atoms
> show sel atoms
> style sel stick
Changed 230 atom styles
> style sel sphere
Changed 230 atom styles
> style sel ball
Changed 230 atom styles
> color sel byhetero
> select clear
> undo
[Repeated 2 time(s)]
> select clear
> save
> D:/Zhou_Lab/Projects/HCMV_Tegument/Figures/Figure_3/UL77_interactions_cleft.cxs
——— End of log from Wed Jun 8 13:51:01 2022 ———
opened ChimeraX session
> ui tool show "Side View"
> view list
Named views: close1, close2, far1, far2
> color #14/E:610,614,618,221,224,225,227,228,530,531,532,534-536,562-567
> #8fe2c1
> color #13/E:251,252,205,206,209,210,466,469 crimson
> select #14/E:610,614,618,221,224,225,227,228,530,531,532,534-536,562-567
164 atoms, 158 bonds, 20 residues, 1 model selected
> select add #13/E:251,252,205,206,209,210,466,469
230 atoms, 219 bonds, 28 residues, 2 models selected
> style sel sphere
Changed 230 atom styles
> style sel ball
Changed 230 atom styles
> style sel sphere
Changed 230 atom styles
> select clear
> view name close3
> view list
Named views: close1, close2, close3, far1, far2
> save C:\Users\jihma\Desktop\image27.png supersample 3
> select #14/E:610,614,618,221,224,225,227,228,530,531,532,534-536,562-567
164 atoms, 158 bonds, 20 residues, 1 model selected
> select add #13/E:251,252,205,206,209,210,466,469
230 atoms, 219 bonds, 28 residues, 2 models selected
> style sel ball
Changed 230 atom styles
> select clear
> undo
[Repeated 2 time(s)]
> color #14/E:610,614,618,221,224,225,227,228,530,531,532,534-536,562-567
> orange
> color #13/E:251,252,205,206,209,210,466,469 lawn green
> select clear
> save C:\Users\jihma\Desktop\image28.png supersample 3
> select #14/E:610,614,618,221,224,225,227,228,530,531,532,534-536,562-567
164 atoms, 158 bonds, 20 residues, 1 model selected
> select add #13/E:251,252,205,206,209,210,466,469
230 atoms, 219 bonds, 28 residues, 2 models selected
> style sel ball
Changed 230 atom styles
> select clear
> save C:\Users\jihma\Desktop\image29.png supersample 3
> save
> D:/Zhou_Lab/Projects/HCMV_Tegument/Figures/Figure_3/UL77_interactions_cleft.cxs
——— End of log from Wed Jun 8 13:59:03 2022 ———
opened ChimeraX session
Traceback (most recent call last):
File "C:\Program Files\ChimeraX 1.4\bin\lib\site-
packages\chimerax\map\volume_viewer.py", line 1530, in <lambda>
QTimer.singleShot(200, lambda f=f: f.setMinimumHeight(50))
RuntimeError: wrapped C/C++ object of type QScrollArea has been deleted
RuntimeError: wrapped C/C++ object of type QScrollArea has been deleted
File "C:\Program Files\ChimeraX 1.4\bin\lib\site-
packages\chimerax\map\volume_viewer.py", line 1530, in
QTimer.singleShot(200, lambda f=f: f.setMinimumHeight(50))
See log for complete Python traceback.
> save
> D:/Zhou_Lab/Projects/HCMV_Tegument/Figures/Figure_3/UL77_interactions_cleft.cxs
——— End of log from Wed Jun 15 14:27:37 2022 ———
opened ChimeraX session
Traceback (most recent call last):
File "C:\Program Files\ChimeraX 1.4\bin\lib\site-
packages\chimerax\map\volume_viewer.py", line 1530, in <lambda>
QTimer.singleShot(200, lambda f=f: f.setMinimumHeight(50))
RuntimeError: wrapped C/C++ object of type QScrollArea has been deleted
RuntimeError: wrapped C/C++ object of type QScrollArea has been deleted
File "C:\Program Files\ChimeraX 1.4\bin\lib\site-
packages\chimerax\map\volume_viewer.py", line 1530, in
QTimer.singleShot(200, lambda f=f: f.setMinimumHeight(50))
See log for complete Python traceback.
OpenGL version: 3.3.0 NVIDIA 512.77
OpenGL renderer: NVIDIA GeForce RTX 3080 Laptop GPU/PCIe/SSE2
OpenGL vendor: NVIDIA Corporation
Python: 3.9.11
Locale: en_US.cp1252
Qt version: PyQt6 6.3.0, Qt 6.3.0
Qt runtime version: 6.3.0
Qt platform: windows
Manufacturer: LENOVO
Model: 82N6
OS: Microsoft Windows 11 Home (Build 22000)
Memory: 34,204,610,560
MaxProcessMemory: 137,438,953,344
CPU: 16 AMD Ryzen 9 5900HX with Radeon Graphics
OSLanguage: en-US
Installed Packages:
alabaster: 0.7.12
appdirs: 1.4.4
Babel: 2.10.1
backcall: 0.2.0
blockdiag: 3.0.0
certifi: 2022.5.18.1
cftime: 1.6.0
charset-normalizer: 2.0.12
ChimeraX-AddCharge: 1.2.3
ChimeraX-AddH: 2.1.11
ChimeraX-AlignmentAlgorithms: 2.0
ChimeraX-AlignmentHdrs: 3.2.1
ChimeraX-AlignmentMatrices: 2.0
ChimeraX-Alignments: 2.4.3
ChimeraX-AlphaFold: 1.0
ChimeraX-AltlocExplorer: 1.0.2
ChimeraX-AmberInfo: 1.0
ChimeraX-Arrays: 1.0
ChimeraX-Atomic: 1.39.1
ChimeraX-AtomicLibrary: 7.0
ChimeraX-AtomSearch: 2.0.1
ChimeraX-AxesPlanes: 2.1
ChimeraX-BasicActions: 1.1
ChimeraX-BILD: 1.0
ChimeraX-BlastProtein: 2.1.1
ChimeraX-BondRot: 2.0
ChimeraX-BugReporter: 1.0
ChimeraX-BuildStructure: 2.7
ChimeraX-Bumps: 1.0
ChimeraX-BundleBuilder: 1.1
ChimeraX-ButtonPanel: 1.0
ChimeraX-CageBuilder: 1.0
ChimeraX-CellPack: 1.0
ChimeraX-Centroids: 1.2
ChimeraX-ChemGroup: 2.0
ChimeraX-Clashes: 2.2.4
ChimeraX-ColorActions: 1.0
ChimeraX-ColorGlobe: 1.0
ChimeraX-ColorKey: 1.5.1
ChimeraX-CommandLine: 1.2.3
ChimeraX-ConnectStructure: 2.0.1
ChimeraX-Contacts: 1.0
ChimeraX-Core: 1.4
ChimeraX-CoreFormats: 1.1
ChimeraX-coulombic: 1.3.2
ChimeraX-Crosslinks: 1.0
ChimeraX-Crystal: 1.0
ChimeraX-CrystalContacts: 1.0
ChimeraX-DataFormats: 1.2.2
ChimeraX-Dicom: 1.1
ChimeraX-DistMonitor: 1.1.5
ChimeraX-Dssp: 2.0
ChimeraX-EMDB-SFF: 1.0
ChimeraX-ExperimentalCommands: 1.0
ChimeraX-FileHistory: 1.0
ChimeraX-FunctionKey: 1.0
ChimeraX-Geometry: 1.2
ChimeraX-gltf: 1.0
ChimeraX-Graphics: 1.1
ChimeraX-Hbonds: 2.1.2
ChimeraX-Help: 1.2
ChimeraX-HKCage: 1.3
ChimeraX-IHM: 1.1
ChimeraX-ImageFormats: 1.2
ChimeraX-IMOD: 1.0
ChimeraX-IO: 1.0.1
ChimeraX-ItemsInspection: 1.0
ChimeraX-Label: 1.1.1
ChimeraX-ListInfo: 1.1.1
ChimeraX-Log: 1.1.5
ChimeraX-LookingGlass: 1.1
ChimeraX-Maestro: 1.8.1
ChimeraX-Map: 1.1
ChimeraX-MapData: 2.0
ChimeraX-MapEraser: 1.0
ChimeraX-MapFilter: 2.0
ChimeraX-MapFit: 2.0
ChimeraX-MapSeries: 2.1
ChimeraX-Markers: 1.0
ChimeraX-Mask: 1.0
ChimeraX-MatchMaker: 2.0.6
ChimeraX-MDcrds: 2.6
ChimeraX-MedicalToolbar: 1.0.1
ChimeraX-Meeting: 1.0
ChimeraX-MLP: 1.1
ChimeraX-mmCIF: 2.7
ChimeraX-MMTF: 2.1
ChimeraX-Modeller: 1.5.5
ChimeraX-ModelPanel: 1.3.2
ChimeraX-ModelSeries: 1.0
ChimeraX-Mol2: 2.0
ChimeraX-Morph: 1.0
ChimeraX-MouseModes: 1.1
ChimeraX-Movie: 1.0
ChimeraX-Neuron: 1.0
ChimeraX-Nucleotides: 2.0.2
ChimeraX-OpenCommand: 1.9
ChimeraX-PDB: 2.6.6
ChimeraX-PDBBio: 1.0
ChimeraX-PDBLibrary: 1.0.2
ChimeraX-PDBMatrices: 1.0
ChimeraX-PickBlobs: 1.0
ChimeraX-Positions: 1.0
ChimeraX-PresetMgr: 1.1
ChimeraX-PubChem: 2.1
ChimeraX-ReadPbonds: 1.0.1
ChimeraX-Registration: 1.1
ChimeraX-RemoteControl: 1.0
ChimeraX-ResidueFit: 1.0
ChimeraX-RestServer: 1.1
ChimeraX-RNALayout: 1.0
ChimeraX-RotamerLibMgr: 2.0.1
ChimeraX-RotamerLibsDunbrack: 2.0
ChimeraX-RotamerLibsDynameomics: 2.0
ChimeraX-RotamerLibsRichardson: 2.0
ChimeraX-SaveCommand: 1.5.1
ChimeraX-SchemeMgr: 1.0
ChimeraX-SDF: 2.0
ChimeraX-Segger: 1.0
ChimeraX-Segment: 1.0
ChimeraX-SelInspector: 1.0
ChimeraX-SeqView: 2.6
ChimeraX-Shape: 1.0.1
ChimeraX-Shell: 1.0
ChimeraX-Shortcuts: 1.1
ChimeraX-ShowAttr: 1.0
ChimeraX-ShowSequences: 1.0
ChimeraX-SideView: 1.0
ChimeraX-Smiles: 2.1
ChimeraX-SmoothLines: 1.0
ChimeraX-SpaceNavigator: 1.0
ChimeraX-StdCommands: 1.8
ChimeraX-STL: 1.0
ChimeraX-Storm: 1.0
ChimeraX-StructMeasure: 1.0.1
ChimeraX-Struts: 1.0.1
ChimeraX-Surface: 1.0
ChimeraX-SwapAA: 2.0
ChimeraX-SwapRes: 2.1.1
ChimeraX-TapeMeasure: 1.0
ChimeraX-Test: 1.0
ChimeraX-Toolbar: 1.1.1
ChimeraX-ToolshedUtils: 1.2.1
ChimeraX-Tug: 1.0
ChimeraX-UI: 1.18.3
ChimeraX-uniprot: 2.2
ChimeraX-UnitCell: 1.0
ChimeraX-ViewDockX: 1.1.2
ChimeraX-VIPERdb: 1.0
ChimeraX-Vive: 1.1
ChimeraX-VolumeMenu: 1.0
ChimeraX-VTK: 1.0
ChimeraX-WavefrontOBJ: 1.0
ChimeraX-WebCam: 1.0
ChimeraX-WebServices: 1.1.0
ChimeraX-Zone: 1.0
colorama: 0.4.4
comtypes: 1.1.10
cxservices: 1.2
cycler: 0.11.0
Cython: 0.29.26
debugpy: 1.6.0
decorator: 5.1.1
docutils: 0.17.1
entrypoints: 0.4
filelock: 3.4.2
fonttools: 4.33.3
funcparserlib: 1.0.0
grako: 3.16.5
h5py: 3.7.0
html2text: 2020.1.16
idna: 3.3
ihm: 0.27
imagecodecs: 2021.11.20
imagesize: 1.3.0
ipykernel: 6.6.1
ipython: 7.31.1
ipython-genutils: 0.2.0
jedi: 0.18.1
Jinja2: 3.0.3
jupyter-client: 7.1.0
jupyter-core: 4.10.0
kiwisolver: 1.4.2
line-profiler: 3.4.0
lxml: 4.7.1
lz4: 3.1.10
MarkupSafe: 2.1.1
matplotlib: 3.5.1
matplotlib-inline: 0.1.3
msgpack: 1.0.3
nest-asyncio: 1.5.5
netCDF4: 1.5.8
networkx: 2.6.3
numexpr: 2.8.1
numpy: 1.22.1
openvr: 1.16.802
packaging: 21.3
ParmEd: 3.4.3
parso: 0.8.3
pickleshare: 0.7.5
Pillow: 9.0.1
pip: 21.3.1
pkginfo: 1.8.2
prompt-toolkit: 3.0.29
psutil: 5.9.0
pycollada: 0.7.2
pydicom: 2.2.2
Pygments: 2.11.2
PyOpenGL: 3.1.5
PyOpenGL-accelerate: 3.1.5
pyparsing: 3.0.9
PyQt6-commercial: 6.3.0
PyQt6-Qt6: 6.3.0
PyQt6-sip: 13.3.1
PyQt6-WebEngine-commercial: 6.3.0
PyQt6-WebEngine-Qt6: 6.3.0
python-dateutil: 2.8.2
pytz: 2022.1
pywin32: 303
pyzmq: 23.1.0
qtconsole: 5.3.0
QtPy: 2.1.0
RandomWords: 0.3.0
requests: 2.27.1
scipy: 1.7.3
setuptools: 59.8.0
sfftk-rw: 0.7.2
six: 1.16.0
snowballstemmer: 2.2.0
sortedcontainers: 2.4.0
Sphinx: 4.3.2
sphinx-autodoc-typehints: 1.15.2
sphinxcontrib-applehelp: 1.0.2
sphinxcontrib-blockdiag: 3.0.0
sphinxcontrib-devhelp: 1.0.2
sphinxcontrib-htmlhelp: 2.0.0
sphinxcontrib-jsmath: 1.0.1
sphinxcontrib-qthelp: 1.0.3
sphinxcontrib-serializinghtml: 1.1.5
suds-community: 1.0.0
tables: 3.7.0
tifffile: 2021.11.2
tinyarray: 1.2.4
tornado: 6.1
traitlets: 5.1.1
urllib3: 1.26.9
wcwidth: 0.2.5
webcolors: 1.11.1
wheel: 0.37.1
wheel-filename: 1.3.0
WMI: 1.5.1
Attachments (2)
Change History (9)
comment:1 by , 4 years ago
| Cc: | added |
|---|---|
| Component: | Unassigned → Sessions |
| Owner: | set to |
| Platform: | → all |
| Project: | → ChimeraX |
| Reporter: | changed from to |
| Status: | new → assigned |
| Summary: | ChimeraX bug report submission → Volume Viewer after session restore: wrapped C/C++ object of type QScrollArea has been deleted |
comment:3 by , 4 years ago
Do your session files include density maps? I don't see anything about density maps in the log of your bug report and yet the error is coming from the Volume Viewer panel that shows density map histograms and controls.
This error message happens because the Volume Viewer panel is created and then destroyed before a timer goes off 0.2 seconds later which is trying to resize the Volume Viewer panel.
I test sessions with and without maps, with and without the volume viewer panel, and they all worked on Mac in ChimeraX 1.4.
Can you provide an example session file that gives this error? Don't send private data since the bug reports are public.
comment:4 by , 4 years ago
I have an idea how this error might happen with the right preference settings, but still it does not happen on a Mac, but maybe it would happen on Windows. Here's the idea. If you have enabled ChimeraX setting Preferences / Window / Resize Graphics Window on Session Restore, and the Volume Viewer panel is shown when you open a session (maybe you auto-start Volume Viewer?) then opening a new session may resize the Chimera window and also it will delete the current volume viewer panel. The error you see is because the volume viewer panel gets a resize notification and then is deleted right after. When a session is restored I would guess that the volume viewer panel is deleted and later the window is resized. But maybe the order gets reversed because I know Qt can delay deleting windows.
At any rate a test of this theory would be to turn off the setting Preferences / Window / Resize Graphics Window on Session Restore and then try opening a session and see if you get the error. Or another test would be to make sure the volume viewer panel is closed before you open a session and see if you get the error.
In any case, I have put a fix in the ChimeraX daily build that checks during the resize notification to make sure the panel has not been closed, so tomorrow's ChimeraX daily build (June 16 2022) should also fix the problem.
comment:5 by , 4 years ago
| Resolution: | → fixed |
|---|---|
| Status: | assigned → closed |
Fixed.
Made code ignore volume viewer resize callback if volume viewer panel has been destroyed.
follow-up: 6 comment:6 by , 4 years ago
Hi Tom,
Thanks for the prompt follow up. I do have Log, Models, Volume Viewer, and
Side View launching by default on startup, which I remember setting as
default in ChimeraX 1.3, and which has seemingly carried over to v1.4
without me having had to do anything. This is what my side panel toolbar
looks like if I just launch ChimeraX 1.4 without any files:
[image: Screenshot_default_ChimX_launch.png]
(The "ui" commands running on default were also previously carried over
from my v1.3.)
However, my "Resize Graphics Window on Session Restore" box under Settings
is not ticked. I also tried your suggestions in all combinations ("Resize
Graphics Windows on Session Restore" ticked/unticked, with maps/without
maps, and Volume Viewer open/closed prior to relaunching a new session) and
the error always pops up. Not that it actually affects the content in my
sessions, buyt in any case, I'll be sure to check the Daily Build tomorrow!
I also noticed a couple other misc items:
1. Although my Side View was set to launch by default in v1.3 and still
does in v1.4 when launching a blank session of ChimeraX, Side View does not
launch at all when restoring a previously saved session in v1.4, regardless
of whether or not Side View was saved or left open when saving the session.
Again, this also isn't a huge issue as I can just reopen Side View, but
thought I'd let you guys know.
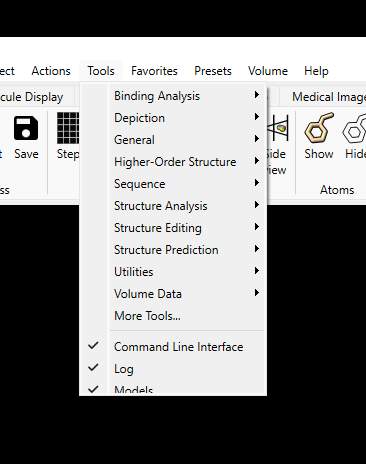
2. A more impactful issue is there also appears to be some drop-down
menu/right click menu UI bugs with v1.4. Upon first launching a blank or
restored session, drop down/right click menus are truncated (please see
image below). On the very top toolbar (File / Edit / Select etc.) this can
be remedied by either reclicking the menu item or hovering over to an
adjacent tab before hovering back. On the dockable toolbar elements, there
seems to be no way to get the full length menu to show. For someone like me
who isn't familiar with all the (great) features you guys have added, this
makes hunting features/exploring a bit difficult.
[image: Screenshot_dropdownmenu_ChimX.png]
Anyhow, appreciate all the effort in this!
Best,
Jonathan
On Wed, Jun 15, 2022 at 5:30 PM ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu>
wrote:
comment:7 by , 4 years ago
Hi Jonathan, Thanks for the details and new bugs. I suspect the 3 problems you describe are all related to our change from the Qt window toolkit version 5 (in ChimeraX 1.3) to the version 6 in ChimeraX 1.4. Also I suspect all 3 problems are specific to Windows, possibly even specific to Windows 11. If you turn off auto-start for Volume Viewer, then restart ChimeraX and open a session I don't think the error should appear. That error can only happen if the volume viewer panel is opened and then closed (and the session file open is what causes it to close). At any rate, tomorrow's daily build will detect the unexpected window resize after deleting the panel and I am pretty sure will not give an error. I will make two new bug reports for the side view and truncated menu problems. Also I will test those on my Windows 10 system. I can test on Windows 11 when I go to the office where we have a Windows 11 machine. Tom

Also, Windows 11 not detected correctly by Bug Reporter