Opened 4 years ago
Closed 3 years ago
#6646 closed enhancement (fixed)
RFE: show modality of DICOM (medical imaging) data
| Reported by: | Elaine Meng | Owned by: | Zach Pearson |
|---|---|---|---|
| Priority: | moderate | Milestone: | |
| Component: | DICOM | Version: | |
| Keywords: | Cc: | Tom Goddard | |
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
It would be useful to make the modality information for each DICOM series more discoverable/obvious, so that it isn't necessary to use some other program like SLICER to understand the contents of datasets from repositories. Of course somebody viewing their own generated data knows the modality, but as soon as there is dataset sharing, this becomes more important. Modality is one of the most prominent aspects of each series (which generally becomes a volume model in ChimeraX).
Here is a page listing DICOM modality attributes
<https://www.dicomlibrary.com/dicom/modality/>
Ideas are to do one or more of the following:
- report series modalit(ies) in the Log when opening DICOM. Remember that the DICOM hierarchy is (1) Patient (2) Study (3) Series, and as I understand it, each series becomes a 3D volume model or 4D volume series in ChimeraX.
- does the level in the DICOM hierarchy also need to be a model attribute? namely patient/study/series
- Prepend the modality when generating the attribute "Volume name" at the series level... that way it would automatically be shown in the Model Panel, reported with "info model", etc.
#1 series0079-Body
1.1 Patient
1.1.1 20220225
1.1.1.1 MR: 785B_4Dflow_ePAT3_retro_native 79
[...]
#1 RIDER-112916-4940
1.1 Patient RIDER-11291640
1.1.1 20060920
1.1.1.1 CT: Chest CT 4
1.1.1.2 SEG: QIN CT challenge: alg01 run2segmentation result 1000
[...]
- also assign modality as a separate volume model attribute which could be reported with "info models," used in command-line specifiers, etc.
Attachments (8)
Change History (36)
comment:2 by , 4 years ago
I strongly suggest not subclassing Volume. This will get you into a nightmare of extra code to try to support session saving. And just in general it does not make sense to subclass to add an attribute.
Elaine's exact suggestions are good ones. Display the modality in the log when the dicom reader opens the data. And have the DICOM reader prepend the modality to the volume name. No new attributes of the Volume instance are needed for either of these, although I agree it may be useful for some future use to have some kind of attribute giving the dicom modality. That could be added to the DICOM grid data class. But then you will want to handle session saving and this gets into a lot of work that you should have a strong reason before you get into that. (It may be useful in the future is not a strong reason.)
The info command is almost never used. While enhancing it might be of some small benefit to us developers, I would not focus on that.
follow-up: 3 comment:3 by , 4 years ago
Why I thought an attribute might be needed is for differential handling of MR vs. CT vs. SEG or RTSTRUCT with regard to: (1) initial display settings, e.g. SEG and RTSTRUCT could automatically be shown as mesh or surface, MR and CT as image style (2) how presets are applied, e.g. whether clicking some icon to adjust thresholds/styles for lung, brain, etc. should only apply to MR and CT, not SEG and RTSTRUCT (keep them as mesh or surface) However, I defer to you as to how important any of this is and even if something we do want to implement, whether it could be accomplished in some other way.
comment:4 by , 4 years ago
Objects should know themselves better than procedural wrappers using those objects know them -- subclassing is a very powerful tool.
follow-up: 6 comment:5 by , 4 years ago
Nothing wrong with having an attribute for modality. As I said subclassing will lead to problems, but if you want to go there and find out for yourself you can. For initial display, the general ChimeraX volume is not going to know anything about DICOM modality, so if you want special initial display when that data is opened then the DICOM module should do it. In general we don't have toolbar icons do different things based on different types of volumes -- that creates behavior the user will have a hard time grasping.
In general I would suggest sticking to the goal of the ticket to make it easy for the user to see the DICOM modality. Implement that in the simplest way. If later issues come up about how to customize the initial display, or how to make buttons recognized the modality of an image, then change the code to support those needs. Keep the code as simple as possible to meet the current needs. If you try to make the code from the start handle imagined future needs it just doesn't work out well.
comment:6 by , 4 years ago
I just get the feeling that you're arguing beyond the scope of what I proposed. No one mentioned changing the behavior of the toolbar or its buttons, I'm not sure where that came from. Subclasses should generally extend an existing API, not override it in a breaking way. There's nothing I have in my head that's in any way related to the toolbar.
I agree, we should do the simplest thing that gets the job done. However, you would never add an outlet to your house by putting bare copper wires in all the terminals of an existing outlet and connecting them to the back of a new outlet located where you wanted your new power source to be. What a fire hazard it would be. So there is a balance between simple and good. I understand it's our job to make that determination. I think you often worry I will go farther than I actually intend to.
comment:7 by , 4 years ago
I see I thought you wrote comment 3 about initial display settings and clicking icons but Elaine wrote that.
Please don't take my comments so seriously.
comment:8 by , 4 years ago
Names on Trac are small :) Heck, I glossed over it entirely, which is why I made the wrong claim nobody mentioned toolbar improvements.
I think it's pretty hard to judge the force of a statement from text, sorry about that.
comment:9 by , 4 years ago
So... it looks like when we open a DICOM file that has a modality, that modality is already shown in the models panel and in the log.
Opening Jason's 4D ultrasound dataset:
Opened US None as #1.1.1.1, grid size 256,176,19, pixel 1, shown at step 1, values uint8
and running info models:
model id #1 type Model name 4D_Ultrasound.dcm model id #1.1 type Model name "Patient " model id #1.1.1 type Model name 20211213 model id #1.1.1.1 type Volume name "US None" model id #1.1.1.1.1 type VolumeImage name image
I think this happens in the Series object's name method.
comment:10 by , 4 years ago
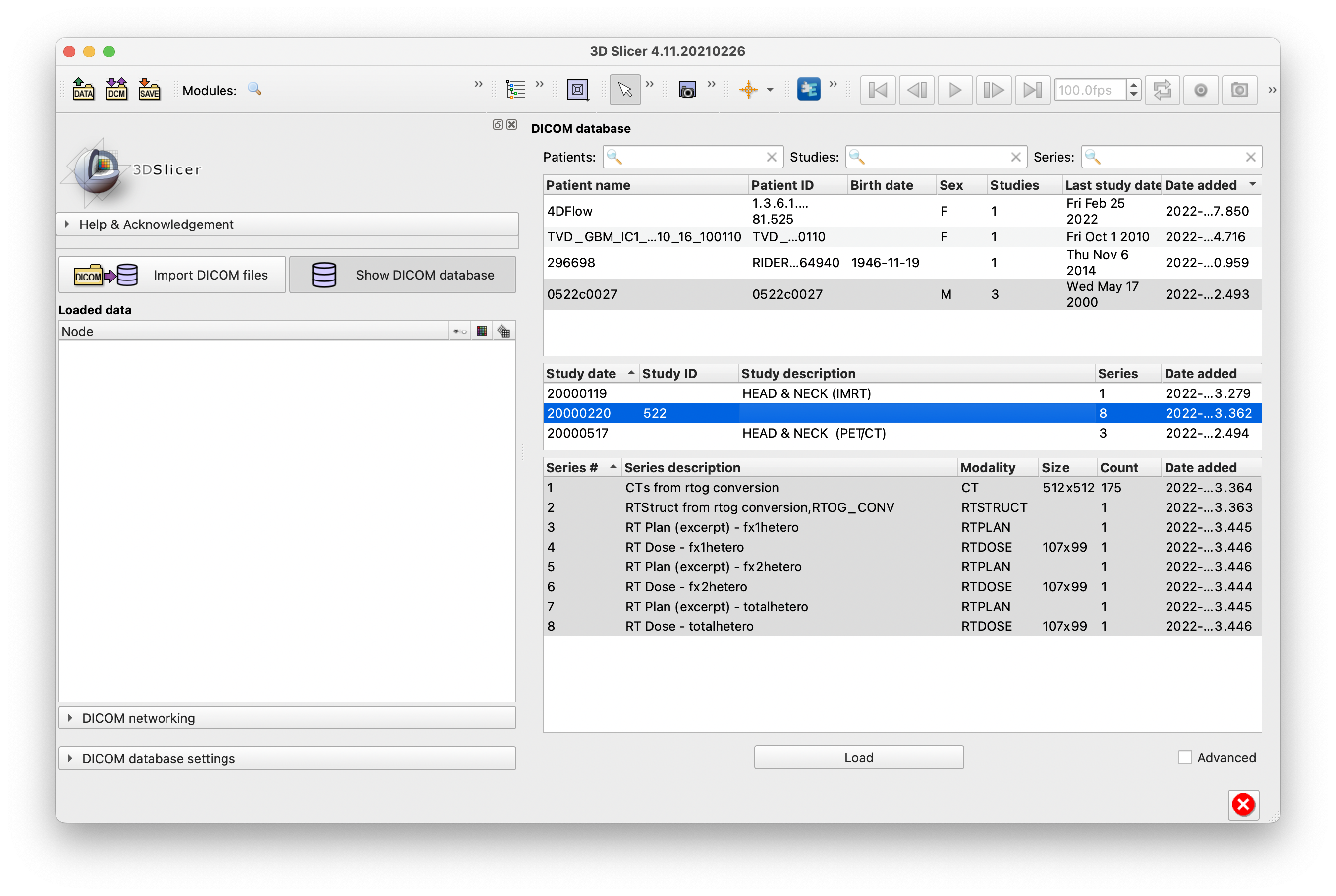
Do you mean the modality is given by the name of model #1 "4D_Ultrasound.dcm"? That appears to come from the file name. I've attached an image of another example of the model names in ChimeraX from Jason. One of the names says 4Dflow not exactly a modality but close.
I think Elaine had in mind modalities like "MR" (magnetic resonance imaging), "CT" (computed tomography), "SEG" (segmentation) and not to rely on those being given in the file names or study names.
by , 4 years ago
| Attachment: | Screen Shot 2022-04-28 at 1.48.18 PM.2.png added |
|---|
Model panel showing model names for Jason Rubenstein data set.
comment:11 by , 4 years ago
No. I believe the "US" in "Volume name "US None"" and in "Opened US None as #1.1.1.1..." refers to the US (Ultrasound) modality code.
comment:12 by , 4 years ago
Oh, got it. I didn't realize was US means. But Jason's first data set does not have "US" in the model names, so I don't think ChimeraX is adding the modality to the model name.
follow-up: 12 comment:13 by , 4 years ago
My other examples don't necessarily have modality in the name. Sometimes the name happens to include the modality. In the RIDER Lung example shown in group meeting and in screenshot here (and included in the example files on plato), both the CT and the associated SEG models include CT in their names, orthogonal to whether that is really their modality. The SEG models also include "segmentation" in their names, but there is no requirement for the name to reveal the modality.
follow-up: 13 comment:14 by , 4 years ago
| Component: | Volume Data → DICOM |
|---|
comment:15 by , 4 years ago
Modality is a specific field in DICOM data -- see the URL in the original text of the ticket. So it should be easy to parse, and at least some of my original suggestions like logging should be low-hanging fruit. Is there anything else I can provide to move this along?
comment:16 by , 3 years ago
We definitely already show the modality when we open models, but only if it's present in the file.
There's not an official "Unknown" tag for modalities. We could use UK, which is not listed in the spec, and warn the user on open: "Series with unknown modalities noted as modality UK", to keep things consistent.
What do you think would be the best UX, Elaine?
comment:17 by , 3 years ago
My suggestions are in the original ticket (before the comments). Namely the following but the middle one was a question, so I guess just the first and third: Ideas are to do one or more of the following: • report series modalit(ies) in the Log when opening DICOM. Remember that the DICOM hierarchy is (1) Patient (2) Study (3) Series, and as I understand it, each series becomes a 3D volume model or 4D volume series in ChimeraX. • does the level in the DICOM hierarchy also need to be a model attribute? namely patient/study/series • Prepend the modality when generating the attribute "Volume name" at the series level... that way it would automatically be shown in the Model Panel, reported with "info model", etc.
comment:18 by , 3 years ago
Let's go with the last one. If you change your mind later let me know. I like "{Modality} Series ({Body Part Examined})" and other information can be shown in the new GUI.
follow-up: 17 comment:19 by , 3 years ago
I think both the first and the third would be valuable. We're not limited to only one or the other, right?
by , 3 years ago
| Attachment: | Screenshot 2022-08-31 at 13.44.14.png added |
|---|
comment:20 by , 3 years ago
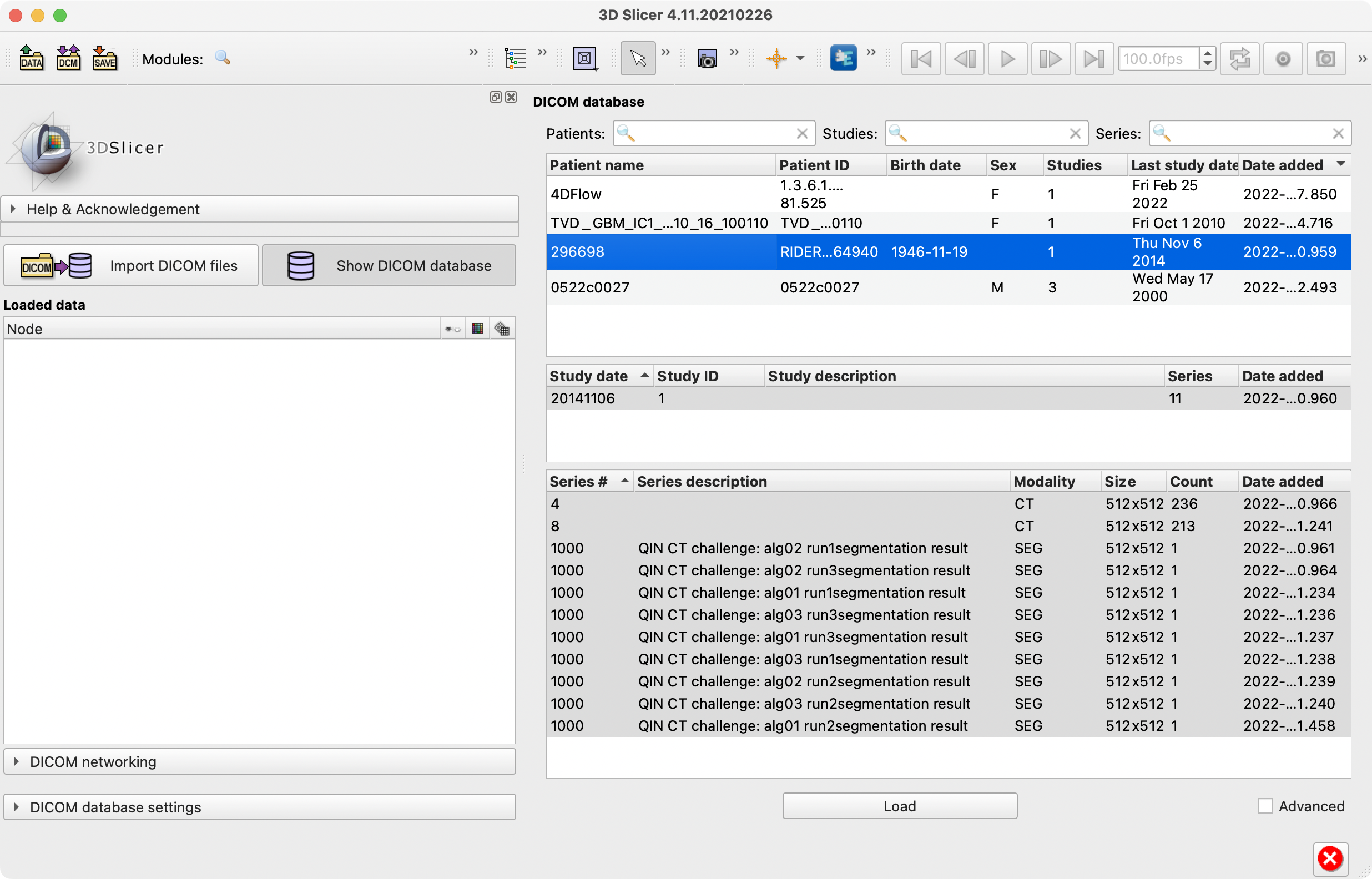
We're not, I just think the first option might get cluttered. I just attached a screenshot of what I think Option 3 should look like. Please ignore the 4 duplicate studies at the end of the hierarchy; I need to figure out why they're being kept alive after merging studies by date.
follow-up: 19 comment:21 by , 3 years ago
It actually looks like Option 1 happens automatically.
Opened CT Series (HEADNECK) as #1.1.2, grid size 512,512,175, pixel 0.98, shown at step 1, values float32 Opened RTDOSE Series (HEADNECK) as #1.1.3, grid size 107,99,118, pixel 3, shown at step 1, values uint16 Opened RTDOSE Series (HEADNECK) as #1.1.4, grid size 107,99,118, pixel 3, shown at step 1, values uint16 Opened RTDOSE Series (HEADNECK) as #1.1.5, grid size 107,99,118, pixel 3, shown at step 1, values uint16
by , 3 years ago
| Attachment: | Screenshot 2022-08-31 at 14.23.09.png added |
|---|
comment:22 by , 3 years ago
Added another example screenshot for the RIDER Lung CT you had posted earlier. I don't see the second series (#8) in either of the examples on Plato, but I think I'm happy with the log and the model hierarchy.
comment:23 by , 3 years ago
My vote is to do both (log and model names) and if we decide log is too much, could turn it off later.
Personally I think it would be good to log all of the following: patient, study, study date, number of series in study, and for each series, description (if available), modality, and size.
But happy to leave you as the point person with final decision. I don't know how hard things are to implement, etc.
Regarding the screenshot, suggestions for model names are to
(1) somehow give unique names for all the series within a study so you can tell which is which
(2) omit repetitive text, or put it at a higher level
Screenshot contains
Patient (ID: NNNN)
Study (date)
regions (RTStruct from rtog conversion)
CT Series (HEADNECK)
RTDOSE Series (HEADNECK)
RTDOSE Series (HEADNECK)
RTDOSE Series (HEADNECK)
Could it instead be something like the following? Slicer shows some of this info as series description, but I don't know if too hard to extract and use in model name. Or in cases where series description includes modality (sometimes it does, sometimes it does not), maybe could just use series description alone.
Patient (ID: NNNN)
Study (date)
RTSTRUCT from rtog conversion
CT from rtog conversion
RTDOSE fx1hetero
RTDOSE fx2hetero
RTDOSE totalhetero
...or some alternatives...
Patient (ID: NNNN)
Study <study number> (date)
CT <series number>
RTDOSE <series number>
RTDOSE <series number>
RTDOSE <series number>
I have other examples where a single study has more than one CT, which is why I suggest appending series number or some kind of disambiguating info from series description. In the example above (if same data as I showed in group meeting), the study ID number is 522, CT series number is 1, RTDOSE series numbers are 4,6,8. Dunno if they are in the file or whether Slicer generated them.
In my other example, there were two CTs in the same study, with series numbers 4 and 8. However, that one (RIDER lung data) had lots of different SEG series as well and they all had the same series number, 1000. Only the "series description" field as shown in Slicer disambiguated them with algorithm and run number, e.g. "alg02 run3"
Of course you can't put all the DICOM browser info into the model names and model hierarchy, but I would like to aim for goals (1) and (2) above. I don't know if that's feasible with the information in the DICOM + reasonable amount of effort, however.
Elaine
by , 3 years ago
| Attachment: | Screenshot 2022-08-31 at 16.26.01.png added |
|---|
comment:24 by , 3 years ago
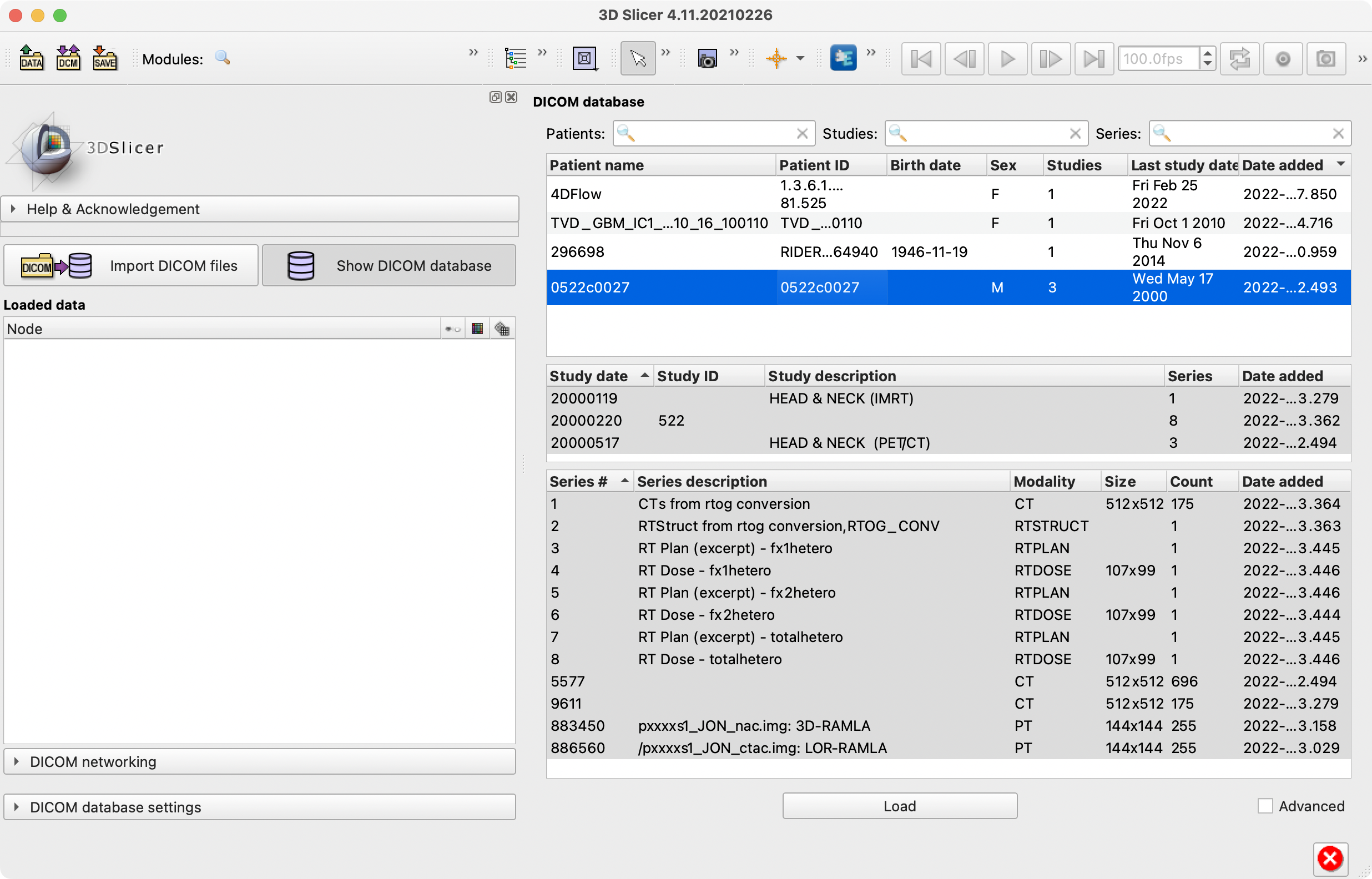
That's way too much to log in my opinion, but how's the naming in this screenshot?
comment:25 by , 3 years ago
Pretty good and much better than previous.
A few more thoughts, but I'm willing to be told it's too hard or wouldn't work well in some cases.
(1) There is some redundancy, namely:
(a) sometimes modality is also in the parenthetical description. Was thinking it could be stripped from that, if present; however, could be difficult and/or turn out weird in other examples.
(b) is body part of the descriptors for a series or for a study? In this example, all of the series in the study are HEADNECK and I'm wondering if it could be put in the study name instead. However, I don't know if a single study might have series in more than one different body part, do you?
(2) could put series in order of their numbers?
Example combining the suggestions:
Patient (ID:NNNN)
Study (date) HEADNECK
1 CT (from rtog conversion)
2 RTSTRUCT (from rtog conversion)
4 RTDOSE (- fx1hetero)
6 RTDOSE (- fx2hetero)
8 RTDOSE (- totalhetero)
comment:26 by , 3 years ago
I don't think it's worth the effort to try to scrub the modalities from the series descriptions.
Body parts are a part of the series metadata. A study can span more than one region, but it's probably obvious what that region is by what's being displayed.
comment:28 by , 3 years ago
| Resolution: | → fixed |
|---|---|
| Status: | assigned → closed |
I think it's a safe assumption that the body part is the same for all series in a study, so I moved it to the study name. If that turns out not to be the case then I'll add a check to ensure that we only put the body part examined in the study name if it's the same for all series.
Series are also now sorted by Series Number under each study.

I like the idea of adding a modality attribute to volume/model objects. I'm thinking of how to make that play nice with the list_info bundle; I'm thinking of subclassing
Volumeand/orModel, adding the attribute, defining__str__, and using that to display info.I think it would also be good to display modalities as you've shown in the examples. Not sure if the hierarchy level needs to be an attribute but I'll look into it.
Those taken together seem like enough for me, but if you think it's too subtle then I can definitely also log modalities.