Opened 5 years ago
Closed 5 years ago
#4238 closed defect (fixed)
Selection outline invisible on segmentations
| Reported by: | Owned by: | Tom Goddard | |
|---|---|---|---|
| Priority: | normal | Milestone: | |
| Component: | Graphics | Version: | |
| Keywords: | Cc: | ||
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
The following bug report has been submitted:
Platform: Darwin-20.2.0-x86_64-i386-64bit
ChimeraX Version: 1.1.1 (2020-10-07 08:32:49 UTC)
Description
Hi, I have segmented a map with Segger and would like to select the segements to group them. With Chimera, I can see a green outline around the segments. In ChimeraX the green outline is invisible. The selection seems to work though. If do the selection blindly, I can group segements. (Describe the actions that caused this problem to occur here)
Log:
UCSF ChimeraX version: 1.1.1 (2020-10-07)
© 2016-2020 Regents of the University of California. All rights reserved.
How to cite UCSF ChimeraX
> open /Volumes/isb-
> data/1_lab_members/Hochheiser_Inga/masks_multibody/octamer.mrc
Opened octamer.mrc, grid size 256,256,256, pixel 1.29, shown at level 0.0152,
step 1, values float32
> volume #1 level 0.01759
> vop gaussian #1 sdev 3.87
> volume #2 level 0.008364
> save
> /Users/gha/Documents/uni/projects/nlrp3_decamer/heidelberg2/octamer_gaussian.mrc
> models #2
> volume #2 level 0.00838
> open
> /Users/gha/Documents/uni/projects/nlrp3_decamer/heidelberg2/MultiBody/run_it008_half1_body001.mrc
Opened run_it008_half1_body001.mrc, grid size 256,256,256, pixel 1.29, shown
at level 0.013, step 1, values float32
> open
> /Users/gha/Documents/uni/projects/nlrp3_decamer/heidelberg2/MultiBody/run_it008_half1_body002.mrc
Opened run_it008_half1_body002.mrc, grid size 256,256,256, pixel 1.29, shown
at level 0.00465, step 1, values float32
> open
> /Users/gha/Documents/uni/projects/nlrp3_decamer/heidelberg2/MultiBody/run_it008_half2_body001.mrc
Opened run_it008_half2_body001.mrc, grid size 256,256,256, pixel 1.29, shown
at level 0.013, step 1, values float32
> open
> /Users/gha/Documents/uni/projects/nlrp3_decamer/heidelberg2/MultiBody/run_it008_half2_body002.mrc
Opened run_it008_half2_body002.mrc, grid size 256,256,256, pixel 1.29, shown
at level 0.00466, step 1, values float32
> volume #4 level 0.01162
> volume #4 level 0.01192
> hide #!2 models
> volume #4 level 0.01118
> volume #6 level 0.01292
> volume #3 level 0.01173
> open
> /Users/gha/Documents/uni/projects/nlrp3_decamer/heidelberg2/3DRefine/run_class001.mrc
Opened run_class001.mrc, grid size 256,256,256, pixel 1.29, shown at level
0.0134, step 1, values float32
> volume #7 level 0.008112
> ui tool show "Segment Map"
Segmenting run_class001.mrc, density threshold 0.008112
Showing run_class001.seg - 0 regions, 0 surfaces
Only showing 60 of 75 regions.
Showing 60 of 75 region surfaces
787 watershed regions, grouped to 75 regions
Segmenting run_class001.mrc, density threshold 0.008112
Showing run_class001.seg - 0 regions, 0 surfaces
Showing 22 region surfaces
787 watershed regions, grouped to 22 regions
Segmenting run_class001.mrc, density threshold 0.008112
Showing run_class001.seg - 0 regions, 0 surfaces
Only showing 60 of 75 regions.
Showing 60 of 75 region surfaces
787 watershed regions, grouped to 75 regions
OpenGL version: 4.1 ATI-4.2.13
OpenGL renderer: AMD Radeon Pro 5500M OpenGL Engine
OpenGL vendor: ATI Technologies Inc.Hardware:
Hardware Overview:
Model Name: MacBook Pro
Model Identifier: MacBookPro16,1
Processor Name: 8-Core Intel Core i9
Processor Speed: 2,4 GHz
Number of Processors: 1
Total Number of Cores: 8
L2 Cache (per Core): 256 KB
L3 Cache: 16 MB
Hyper-Threading Technology: Enabled
Memory: 16 GB
System Firmware Version: 1554.60.15.0.0 (iBridge: 18.16.13030.0.0,0)
Software:
System Software Overview:
System Version: macOS 11.1 (20C69)
Kernel Version: Darwin 20.2.0
Time since boot: 1 day 3:27
Graphics/Displays:
Intel UHD Graphics 630:
Chipset Model: Intel UHD Graphics 630
Type: GPU
Bus: Built-In
VRAM (Dynamic, Max): 1536 MB
Vendor: Intel
Device ID: 0x3e9b
Revision ID: 0x0002
Automatic Graphics Switching: Supported
gMux Version: 5.0.0
Metal Family: Supported, Metal GPUFamily macOS 2
AMD Radeon Pro 5500M:
Chipset Model: AMD Radeon Pro 5500M
Type: GPU
Bus: PCIe
PCIe Lane Width: x8
VRAM (Total): 8 GB
Vendor: AMD (0x1002)
Device ID: 0x7340
Revision ID: 0x0040
ROM Revision: 113-D3220E-190
VBIOS Version: 113-D32206U1-019
Option ROM Version: 113-D32206U1-019
EFI Driver Version: 01.A1.190
Automatic Graphics Switching: Supported
gMux Version: 5.0.0
Metal Family: Supported, Metal GPUFamily macOS 2
Displays:
Color LCD:
Display Type: Built-In Retina LCD
Resolution: 3072 x 1920 Retina
Framebuffer Depth: 30-Bit Color (ARGB2101010)
Mirror: Off
Online: Yes
Automatically Adjust Brightness: Yes
Connection Type: Internal
DELL U3818DW:
Resolution: 3840 x 1600 (Ultra-wide 4K)
UI Looks like: 3840 x 1600 @ 60.00Hz
Framebuffer Depth: 30-Bit Color (ARGB2101010)
Display Serial Number: 5KC038CE0FRL
Main Display: Yes
Mirror: Off
Online: Yes
Rotation: Supported
Automatically Adjust Brightness: No
Connection Type: Thunderbolt/DisplayPort
Television: Yes
PyQt version: 5.12.3
Compiled Qt version: 5.12.4
Runtime Qt version: 5.12.9
Installed Packages:
alabaster: 0.7.12
appdirs: 1.4.4
appnope: 0.1.0
Babel: 2.8.0
backcall: 0.2.0
blockdiag: 2.0.1
certifi: 2020.6.20
chardet: 3.0.4
ChimeraX-AddH: 2.1.3
ChimeraX-AlignmentAlgorithms: 2.0
ChimeraX-AlignmentHdrs: 3.2
ChimeraX-AlignmentMatrices: 2.0
ChimeraX-Alignments: 2.1
ChimeraX-Arrays: 1.0
ChimeraX-Atomic: 1.6.1
ChimeraX-AtomSearch: 2.0
ChimeraX-AxesPlanes: 2.0
ChimeraX-BasicActions: 1.1
ChimeraX-BILD: 1.0
ChimeraX-BlastProtein: 1.0.1
ChimeraX-BondRot: 2.0
ChimeraX-BugReporter: 1.0
ChimeraX-BuildStructure: 2.0
ChimeraX-Bumps: 1.0
ChimeraX-BundleBuilder: 1.0
ChimeraX-ButtonPanel: 1.0
ChimeraX-CageBuilder: 1.0
ChimeraX-CellPack: 1.0
ChimeraX-Centroids: 1.1
ChimeraX-ChemGroup: 2.0
ChimeraX-Clashes: 2.0
ChimeraX-ColorActions: 1.0
ChimeraX-ColorGlobe: 1.0
ChimeraX-CommandLine: 1.1.3
ChimeraX-ConnectStructure: 2.0
ChimeraX-Contacts: 1.0
ChimeraX-Core: 1.1.1
ChimeraX-CoreFormats: 1.0
ChimeraX-coulombic: 1.0.1
ChimeraX-Crosslinks: 1.0
ChimeraX-Crystal: 1.0
ChimeraX-DataFormats: 1.0
ChimeraX-Dicom: 1.0
ChimeraX-DistMonitor: 1.1
ChimeraX-DistUI: 1.0
ChimeraX-Dssp: 2.0
ChimeraX-EMDB-SFF: 1.0
ChimeraX-ExperimentalCommands: 1.0
ChimeraX-FileHistory: 1.0
ChimeraX-FunctionKey: 1.0
ChimeraX-Geometry: 1.1
ChimeraX-gltf: 1.0
ChimeraX-Graphics: 1.0
ChimeraX-Hbonds: 2.0
ChimeraX-Help: 1.0
ChimeraX-HKCage: 1.3
ChimeraX-IHM: 1.0
ChimeraX-ImageFormats: 1.0
ChimeraX-IMOD: 1.0
ChimeraX-IO: 1.0
ChimeraX-Label: 1.0
ChimeraX-ListInfo: 1.0
ChimeraX-Log: 1.1.1
ChimeraX-LookingGlass: 1.1
ChimeraX-Map: 1.0.1
ChimeraX-MapData: 2.0
ChimeraX-MapEraser: 1.0
ChimeraX-MapFilter: 2.0
ChimeraX-MapFit: 2.0
ChimeraX-MapSeries: 2.0
ChimeraX-Markers: 1.0
ChimeraX-Mask: 1.0
ChimeraX-MatchMaker: 1.1
ChimeraX-MDcrds: 2.0
ChimeraX-MedicalToolbar: 1.0.1
ChimeraX-Meeting: 1.0
ChimeraX-MLP: 1.0
ChimeraX-mmCIF: 2.2
ChimeraX-MMTF: 2.0
ChimeraX-Modeller: 1.0
ChimeraX-ModelPanel: 1.0
ChimeraX-ModelSeries: 1.0
ChimeraX-Mol2: 2.0
ChimeraX-Morph: 1.0
ChimeraX-MouseModes: 1.0
ChimeraX-Movie: 1.0
ChimeraX-Neuron: 1.0
ChimeraX-Nucleotides: 2.0
ChimeraX-OpenCommand: 1.2.1
ChimeraX-PDB: 2.1
ChimeraX-PDBBio: 1.0
ChimeraX-PickBlobs: 1.0
ChimeraX-Positions: 1.0
ChimeraX-PresetMgr: 1.0
ChimeraX-PubChem: 2.0
ChimeraX-Read-Pbonds: 1.0
ChimeraX-Registration: 1.1
ChimeraX-RemoteControl: 1.0
ChimeraX-ResidueFit: 1.0
ChimeraX-RestServer: 1.0
ChimeraX-RNALayout: 1.0
ChimeraX-RotamerLibMgr: 2.0
ChimeraX-RotamerLibsDunbrack: 2.0
ChimeraX-RotamerLibsDynameomics: 2.0
ChimeraX-RotamerLibsRichardson: 2.0
ChimeraX-SaveCommand: 1.2
ChimeraX-SchemeMgr: 1.0
ChimeraX-SDF: 2.0
ChimeraX-Segger: 1.0
ChimeraX-Segment: 1.0
ChimeraX-SeqView: 2.2
ChimeraX-Shape: 1.0.1
ChimeraX-Shell: 1.0
ChimeraX-Shortcuts: 1.0
ChimeraX-ShowAttr: 1.0
ChimeraX-ShowSequences: 1.0
ChimeraX-SideView: 1.0
ChimeraX-Smiles: 2.0
ChimeraX-SmoothLines: 1.0
ChimeraX-SpaceNavigator: 1.0
ChimeraX-StdCommands: 1.0.4
ChimeraX-STL: 1.0
ChimeraX-Storm: 1.0
ChimeraX-Struts: 1.0
ChimeraX-Surface: 1.0
ChimeraX-SwapAA: 2.0
ChimeraX-SwapRes: 2.0
ChimeraX-TapeMeasure: 1.0
ChimeraX-Test: 1.0
ChimeraX-Toolbar: 1.0
ChimeraX-ToolshedUtils: 1.0
ChimeraX-Tug: 1.0
ChimeraX-UI: 1.2.3
ChimeraX-uniprot: 2.0
ChimeraX-ViewDockX: 1.0
ChimeraX-Vive: 1.1
ChimeraX-VolumeMenu: 1.0
ChimeraX-VTK: 1.0
ChimeraX-WavefrontOBJ: 1.0
ChimeraX-WebCam: 1.0
ChimeraX-WebServices: 1.0
ChimeraX-Zone: 1.0
colorama: 0.4.3
comtypes: 1.1.7
cxservices: 1.0
cycler: 0.10.0
Cython: 0.29.20
decorator: 4.4.2
distlib: 0.3.1
docutils: 0.16
filelock: 3.0.12
funcparserlib: 0.3.6
grako: 3.16.5
h5py: 2.10.0
html2text: 2020.1.16
idna: 2.10
ihm: 0.16
imagecodecs: 2020.5.30
imagecodecs-lite: 2020.1.31
imagesize: 1.2.0
ipykernel: 5.3.0
ipython: 7.15.0
ipython-genutils: 0.2.0
jedi: 0.17.2
Jinja2: 2.11.2
jupyter-client: 6.1.3
jupyter-core: 4.6.3
kiwisolver: 1.2.0
line-profiler: 2.1.2
lxml: 4.5.1
MarkupSafe: 1.1.1
matplotlib: 3.2.1
msgpack: 1.0.0
netifaces: 0.10.9
networkx: 2.4
numexpr: 2.7.1
numpy: 1.18.5
numpydoc: 1.0.0
openvr: 1.12.501
packaging: 20.4
parso: 0.7.1
pexpect: 4.8.0
pickleshare: 0.7.5
Pillow: 7.1.2
pip: 20.2.2
pkginfo: 1.5.0.1
prompt-toolkit: 3.0.7
psutil: 5.7.0
ptyprocess: 0.6.0
pycollada: 0.7.1
pydicom: 2.0.0
Pygments: 2.6.1
PyOpenGL: 3.1.5
PyOpenGL-accelerate: 3.1.5
pyparsing: 2.4.7
PyQt5-commercial: 5.12.3
PyQt5-sip: 4.19.19
PyQtWebEngine-commercial: 5.12.1
python-dateutil: 2.8.1
pytz: 2020.1
pyzmq: 19.0.2
qtconsole: 4.7.4
QtPy: 1.9.0
RandomWords: 0.3.0
requests: 2.24.0
scipy: 1.4.1
setuptools: 49.4.0
sfftk-rw: 0.6.6.dev0
six: 1.15.0
snowballstemmer: 2.0.0
sortedcontainers: 2.2.2
Sphinx: 3.1.1
sphinxcontrib-applehelp: 1.0.2
sphinxcontrib-blockdiag: 2.0.0
sphinxcontrib-devhelp: 1.0.2
sphinxcontrib-htmlhelp: 1.0.3
sphinxcontrib-jsmath: 1.0.1
sphinxcontrib-qthelp: 1.0.3
sphinxcontrib-serializinghtml: 1.1.4
suds-jurko: 0.6
tables: 3.6.1
tifffile: 2020.6.3
tinyarray: 1.2.2
tornado: 6.0.4
traitlets: 5.0.4
urllib3: 1.25.10
wcwidth: 0.2.5
webcolors: 1.11.1
wheel: 0.34.2
Attachments (1)
Change History (9)
comment:1 by , 5 years ago
| Component: | Unassigned → Graphics |
|---|---|
| Owner: | set to |
| Platform: | → all |
| Project: | → ChimeraX |
| Status: | new → assigned |
| Summary: | ChimeraX bug report submission → Selection outline invisible on segmentations |
comment:2 by , 5 years ago
This sounds like a macOS graphics driver bug. If that is the problem I would expect no objects show a green selection outline. Is the green outline shown if you you select your MRC map surface?
You can see in the Log panel when you select a volume it shows the equivalent command, e.g. "select #1". It does not show a command for selecting a segmentation segment I guess because there is no command that does that (no way to name the segment).
I tried selecting a segmentation surface (open 1547 from emdb, segment, ctrl-click segment) with the same version of ChimeraX (1.1.1) on macOS 11.2 and the green outline is shown. My machine had different graphics AMD Radeon Pro 580.
comment:3 by , 5 years ago
I see you have two displays. Does the green outline appear if you run ChimeraX without a second display? Maybe the graphics driver has trouble with the external display. Not likely, but worth trying.
follow-up: 4 comment:4 by , 5 years ago
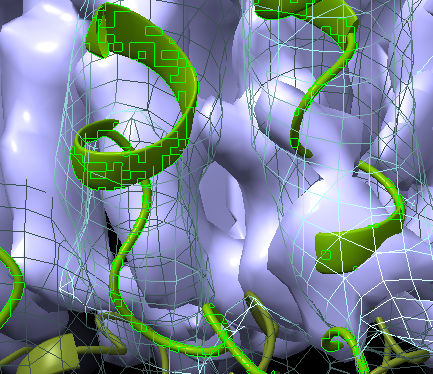
Hi, Thanks for your help! Here is an example when I select a molecule. It also looks weird and the green boxes change all the time when I rotate the view. Cheers, Gregor ----- PD Dr. Gregor Hagelueken Institute of Structural Biology Biomedical Center (BMZ) Building 13, Rm 2G 022/023 University of Bonn Venusberg Campus 1 53127 Bonn, Germany Phone: +49 228 287-51200 WWW: http://www.isb.uni-bonn.de/ ----- On 11. Feb 2021, 23:59 +0100, ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu>, wrote:
comment:5 by , 5 years ago
So selection outlines are messed up. This is probably because the Apple graphics driver is doing something very unusual, giving incompatible offscreen and onscreen framebuffers. The green outline is rendered offscreen and then copied to the screen. It might have to do with differences in multisampling. You can make ChimeraX render everything (models and outlines) offscreen and then copy to the screen with this ChimeraX command
graphics quality colorDepth 16
If you use that command and then select do the outlines appear correct?
comment:6 by , 5 years ago
Hi Tom, Cool, that works! Thanks and have a nice WE, Gregor ----- PD Dr. Gregor Hagelueken Institute of Structural Biology Biomedical Center (BMZ) Building 13, Rm 2G 022/023 University of Bonn Venusberg Campus 1 53127 Bonn, Germany Phone: +49 228 287-51200 WWW: http://www.isb.uni-bonn.de/ ----- On 12. Feb 2021, 20:14 +0100, ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu>, wrote:
follow-up: 6 comment:7 by , 5 years ago
Ok, I have put a fix in so ChimeraX will automatically use the offscreen mode when drawing selection outlines for macOS with AMD Radeon Pro 5500M graphics. So if you use tonight's ChimeraX daily build or newer you will not have to use the colorDepth trick to fix the outlines, they will just work.
follow-up: 7 comment:8 by , 5 years ago
| Resolution: | → fixed |
|---|---|
| Status: | assigned → closed |
Reported by Gregor Hagelueken