Opened 3 years ago
Closed 3 years ago
#8055 closed defect (fixed)
AlphaFold: I/O operation on closed file
| Reported by: | Owned by: | Tom Goddard | |
|---|---|---|---|
| Priority: | normal | Milestone: | |
| Component: | Structure Prediction | Version: | |
| Keywords: | Cc: | ||
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
The following bug report has been submitted:
Platform: macOS-10.16-x86_64-i386-64bit
ChimeraX Version: 1.4 (2022-06-03 23:39:42 UTC)
Description
I'm using Alphafold to predict complexes between 2 proteins. It has worked many times, but the last few times give an error message (see screenshot attached) 'I/O operation on closed file'. I'd appreciate any advice you can give me.
Log:
UCSF ChimeraX version: 1.4 (2022-06-03)
© 2016-2022 Regents of the University of California. All rights reserved.
How to cite UCSF ChimeraX
> ui tool show AlphaFold
> alphafold predict
> MSSDKSNLLKKYKIVFLGDQSVGKTSLITRFMYDTFDETYAATIGIDFLSKTMYLEEGKTIRLQLWDTAGQERFRSLIPSYIRDSHVAVICYDITNKKSFDNLDKWIKDVKLERGDDVIIVLVGNKSDLASDKRQVSLDDVENLQIKIGAKFFIETSTKANHNVKLLFKKIAQSLPDFNQDSNDKSNDNNNNNNNNQSETIDITIDNTAPNPQGTSTCC,MKSDLKTYRSKVLFECKDGASLYSIPCTDNLSTIKFTERNKIWQGSLLFIEEEQLSNSYMEGFEQTKNDKFIIHPFLKMKSKLEFRNGRYLWGETWYCPVVEDLNLSTTTATNVDVQKVTKISHDGTNTIEQISSNKFKVIVQLPNSGYKPYYRNHSPDDLPQVALGLEFENPDMAAQFEYMLAVYDARFKYESQSYYYDRMVYAVNIINFKTIMQEQEEEEEEDIHELTEEFKNQNITDEFSYYEKTSKELEEFEKENANKFDHDTNDDDDGNDDEDDDFGDFIAC
Running AlphaFold prediction
OpenGL version: 4.1 ATI-4.8.101
OpenGL renderer: AMD Radeon Pro 580X OpenGL Engine
OpenGL vendor: ATI Technologies Inc.
Python: 3.9.11
Locale: UTF-8
Qt version: PyQt6 6.3.0, Qt 6.3.0
Qt runtime version: 6.3.0
Qt platform: cocoa
Hardware:
Hardware Overview:
Model Name: iMac
Model Identifier: iMac19,1
Processor Name: 8-Core Intel Core i9
Processor Speed: 3.6 GHz
Number of Processors: 1
Total Number of Cores: 8
L2 Cache (per Core): 256 KB
L3 Cache: 16 MB
Hyper-Threading Technology: Enabled
Memory: 64 GB
System Firmware Version: 1916.40.6.0.0
OS Loader Version: 540.120.3~22
SMC Version (system): 2.46f12
Software:
System Software Overview:
System Version: macOS 12.6.1 (21G217)
Kernel Version: Darwin 21.6.0
Time since boot: 6 days 18:12
Graphics/Displays:
Radeon Pro 580X:
Chipset Model: Radeon Pro 580X
Type: GPU
Bus: PCIe
PCIe Lane Width: x16
VRAM (Total): 8 GB
Vendor: AMD (0x1002)
Device ID: 0x67df
Revision ID: 0x00c0
ROM Revision: 113-D0008A-042
VBIOS Version: 113-D0008A1X-009
EFI Driver Version: 01.B1.042
Metal Family: Supported, Metal GPUFamily macOS 2
Displays:
iMac:
Display Type: Built-In Retina LCD
Resolution: Retina 5K (5120 x 2880)
Framebuffer Depth: 30-Bit Color (ARGB2101010)
Main Display: Yes
Mirror: Off
Online: Yes
Automatically Adjust Brightness: Yes
Connection Type: Internal
Thunderbolt Display:
Display Type: LCD
Resolution: 2048 x 1152
UI Looks like: 2048 x 1152
Framebuffer Depth: 30-Bit Color (ARGB2101010)
Display Serial Number: C02MC33KF2GC
Mirror: Off
Online: Yes
Rotation: Supported
Automatically Adjust Brightness: No
Connection Type: Thunderbolt/DisplayPort
Installed Packages:
alabaster: 0.7.12
appdirs: 1.4.4
appnope: 0.1.3
Babel: 2.10.1
backcall: 0.2.0
blockdiag: 3.0.0
certifi: 2021.10.8
cftime: 1.6.0
charset-normalizer: 2.0.12
ChimeraX-AddCharge: 1.2.3
ChimeraX-AddH: 2.1.11
ChimeraX-AlignmentAlgorithms: 2.0
ChimeraX-AlignmentHdrs: 3.2.1
ChimeraX-AlignmentMatrices: 2.0
ChimeraX-Alignments: 2.4.3
ChimeraX-AlphaFold: 1.0
ChimeraX-AltlocExplorer: 1.0.2
ChimeraX-AmberInfo: 1.0
ChimeraX-Arrays: 1.0
ChimeraX-Atomic: 1.39.1
ChimeraX-AtomicLibrary: 7.0
ChimeraX-AtomSearch: 2.0.1
ChimeraX-AxesPlanes: 2.1
ChimeraX-BasicActions: 1.1
ChimeraX-BILD: 1.0
ChimeraX-BlastProtein: 2.1.1
ChimeraX-BondRot: 2.0
ChimeraX-BugReporter: 1.0
ChimeraX-BuildStructure: 2.7
ChimeraX-Bumps: 1.0
ChimeraX-BundleBuilder: 1.1
ChimeraX-ButtonPanel: 1.0
ChimeraX-CageBuilder: 1.0
ChimeraX-CellPack: 1.0
ChimeraX-Centroids: 1.2
ChimeraX-ChemGroup: 2.0
ChimeraX-Clashes: 2.2.4
ChimeraX-ColorActions: 1.0
ChimeraX-ColorGlobe: 1.0
ChimeraX-ColorKey: 1.5.1
ChimeraX-CommandLine: 1.2.3
ChimeraX-ConnectStructure: 2.0.1
ChimeraX-Contacts: 1.0
ChimeraX-Core: 1.4
ChimeraX-CoreFormats: 1.1
ChimeraX-coulombic: 1.3.2
ChimeraX-Crosslinks: 1.0
ChimeraX-Crystal: 1.0
ChimeraX-CrystalContacts: 1.0
ChimeraX-DataFormats: 1.2.2
ChimeraX-Dicom: 1.1
ChimeraX-DistMonitor: 1.1.5
ChimeraX-Dssp: 2.0
ChimeraX-EMDB-SFF: 1.0
ChimeraX-ExperimentalCommands: 1.0
ChimeraX-FileHistory: 1.0
ChimeraX-FunctionKey: 1.0
ChimeraX-Geometry: 1.2
ChimeraX-gltf: 1.0
ChimeraX-Graphics: 1.1
ChimeraX-Hbonds: 2.1.2
ChimeraX-Help: 1.2
ChimeraX-HKCage: 1.3
ChimeraX-IHM: 1.1
ChimeraX-ImageFormats: 1.2
ChimeraX-IMOD: 1.0
ChimeraX-IO: 1.0.1
ChimeraX-ItemsInspection: 1.0
ChimeraX-Label: 1.1.1
ChimeraX-ListInfo: 1.1.1
ChimeraX-Log: 1.1.5
ChimeraX-LookingGlass: 1.1
ChimeraX-Maestro: 1.8.1
ChimeraX-Map: 1.1
ChimeraX-MapData: 2.0
ChimeraX-MapEraser: 1.0
ChimeraX-MapFilter: 2.0
ChimeraX-MapFit: 2.0
ChimeraX-MapSeries: 2.1
ChimeraX-Markers: 1.0
ChimeraX-Mask: 1.0
ChimeraX-MatchMaker: 2.0.6
ChimeraX-MDcrds: 2.6
ChimeraX-MedicalToolbar: 1.0.1
ChimeraX-Meeting: 1.0
ChimeraX-MLP: 1.1
ChimeraX-mmCIF: 2.7
ChimeraX-MMTF: 2.1
ChimeraX-Modeller: 1.5.5
ChimeraX-ModelPanel: 1.3.2
ChimeraX-ModelSeries: 1.0
ChimeraX-Mol2: 2.0
ChimeraX-Morph: 1.0
ChimeraX-MouseModes: 1.1
ChimeraX-Movie: 1.0
ChimeraX-Neuron: 1.0
ChimeraX-Nucleotides: 2.0.2
ChimeraX-OpenCommand: 1.9
ChimeraX-PDB: 2.6.6
ChimeraX-PDBBio: 1.0
ChimeraX-PDBLibrary: 1.0.2
ChimeraX-PDBMatrices: 1.0
ChimeraX-PickBlobs: 1.0
ChimeraX-Positions: 1.0
ChimeraX-PresetMgr: 1.1
ChimeraX-PubChem: 2.1
ChimeraX-ReadPbonds: 1.0.1
ChimeraX-Registration: 1.1
ChimeraX-RemoteControl: 1.0
ChimeraX-ResidueFit: 1.0
ChimeraX-RestServer: 1.1
ChimeraX-RNALayout: 1.0
ChimeraX-RotamerLibMgr: 2.0.1
ChimeraX-RotamerLibsDunbrack: 2.0
ChimeraX-RotamerLibsDynameomics: 2.0
ChimeraX-RotamerLibsRichardson: 2.0
ChimeraX-SaveCommand: 1.5.1
ChimeraX-SchemeMgr: 1.0
ChimeraX-SDF: 2.0
ChimeraX-Segger: 1.0
ChimeraX-Segment: 1.0
ChimeraX-SelInspector: 1.0
ChimeraX-SeqView: 2.6
ChimeraX-Shape: 1.0.1
ChimeraX-Shell: 1.0
ChimeraX-Shortcuts: 1.1
ChimeraX-ShowAttr: 1.0
ChimeraX-ShowSequences: 1.0
ChimeraX-SideView: 1.0
ChimeraX-Smiles: 2.1
ChimeraX-SmoothLines: 1.0
ChimeraX-SpaceNavigator: 1.0
ChimeraX-StdCommands: 1.8
ChimeraX-STL: 1.0
ChimeraX-Storm: 1.0
ChimeraX-StructMeasure: 1.0.1
ChimeraX-Struts: 1.0.1
ChimeraX-Surface: 1.0
ChimeraX-SwapAA: 2.0
ChimeraX-SwapRes: 2.1.1
ChimeraX-TapeMeasure: 1.0
ChimeraX-Test: 1.0
ChimeraX-Toolbar: 1.1.1
ChimeraX-ToolshedUtils: 1.2.1
ChimeraX-Tug: 1.0
ChimeraX-UI: 1.18.3
ChimeraX-uniprot: 2.2
ChimeraX-UnitCell: 1.0
ChimeraX-ViewDockX: 1.1.2
ChimeraX-VIPERdb: 1.0
ChimeraX-Vive: 1.1
ChimeraX-VolumeMenu: 1.0
ChimeraX-VTK: 1.0
ChimeraX-WavefrontOBJ: 1.0
ChimeraX-WebCam: 1.0
ChimeraX-WebServices: 1.1.0
ChimeraX-Zone: 1.0
colorama: 0.4.4
cxservices: 1.2
cycler: 0.11.0
Cython: 0.29.26
debugpy: 1.6.0
decorator: 5.1.1
docutils: 0.17.1
entrypoints: 0.4
filelock: 3.4.2
fonttools: 4.33.3
funcparserlib: 1.0.0
grako: 3.16.5
h5py: 3.7.0
html2text: 2020.1.16
idna: 3.3
ihm: 0.27
imagecodecs: 2021.11.20
imagesize: 1.3.0
ipykernel: 6.6.1
ipython: 7.31.1
ipython-genutils: 0.2.0
jedi: 0.18.1
Jinja2: 3.0.3
jupyter-client: 7.1.0
jupyter-core: 4.10.0
kiwisolver: 1.4.2
line-profiler: 3.4.0
lxml: 4.7.1
lz4: 3.1.10
MarkupSafe: 2.1.1
matplotlib: 3.5.1
matplotlib-inline: 0.1.3
msgpack: 1.0.3
nest-asyncio: 1.5.5
netCDF4: 1.5.8
networkx: 2.6.3
numexpr: 2.8.1
numpy: 1.22.1
openvr: 1.16.802
packaging: 21.0
ParmEd: 3.4.3
parso: 0.8.3
pexpect: 4.8.0
pickleshare: 0.7.5
Pillow: 9.0.1
pip: 21.3.1
pkginfo: 1.8.2
prompt-toolkit: 3.0.29
psutil: 5.9.0
ptyprocess: 0.7.0
pycollada: 0.7.2
pydicom: 2.2.2
Pygments: 2.11.2
PyOpenGL: 3.1.5
PyOpenGL-accelerate: 3.1.5
pyparsing: 3.0.9
PyQt6-commercial: 6.3.0
PyQt6-Qt6: 6.3.0
PyQt6-sip: 13.3.1
PyQt6-WebEngine-commercial: 6.3.0
PyQt6-WebEngine-Qt6: 6.3.0
python-dateutil: 2.8.2
pytz: 2022.1
pyzmq: 23.1.0
qtconsole: 5.3.0
QtPy: 2.1.0
RandomWords: 0.3.0
requests: 2.27.1
scipy: 1.7.3
setuptools: 59.8.0
sfftk-rw: 0.7.2
six: 1.16.0
snowballstemmer: 2.2.0
sortedcontainers: 2.4.0
Sphinx: 4.3.2
sphinx-autodoc-typehints: 1.15.2
sphinxcontrib-applehelp: 1.0.2
sphinxcontrib-blockdiag: 3.0.0
sphinxcontrib-devhelp: 1.0.2
sphinxcontrib-htmlhelp: 2.0.0
sphinxcontrib-jsmath: 1.0.1
sphinxcontrib-qthelp: 1.0.3
sphinxcontrib-serializinghtml: 1.1.5
suds-community: 1.0.0
tables: 3.7.0
tifffile: 2021.11.2
tinyarray: 1.2.4
tornado: 6.1
traitlets: 5.1.1
urllib3: 1.26.9
wcwidth: 0.2.5
webcolors: 1.11.1
wheel: 0.37.1
wheel-filename: 1.3.0
File attachment: Screen Shot 2022-11-19 at 11.00.32 AM.png
Attachments (7)
Change History (15)
by , 3 years ago
| Attachment: | Screen Shot 2022-11-19 at 11.00.32 AM.png added |
|---|
comment:1 by , 3 years ago
| Component: | Unassigned → Structure Prediction |
|---|---|
| Owner: | set to |
| Platform: | → all |
| Project: | → ChimeraX |
| Status: | new → assigned |
| Summary: | ChimeraX bug report submission → AlphaFold: I/O operation on closed file |
Reported by Bernard Lemire
comment:2 by , 3 years ago
| Resolution: | → nonchimerax |
|---|---|
| Status: | assigned → closed |
That is an error in AlphaFold code, not in ChimeraX. It says it is routine overwrite_b_factors() in line 69 of
alphafold/relax/utils.py
and reports
ValueError: I/O operation on closed file
Looking at that code it is adding b-factor to PDB. It is odd because that code just writes the PDB to a in memory string buffer and returns the value. The input to the function is a string and the output is a string, and I don't see any files being used in that routine. Maybe the error is just misleading. Searching online does suggest that the in memory StringIO will produce this error if it has been closed and then you try to read from it. That doesn't look like it could happen in the AlphaFold code. The preceding line saved the PDB file to this string buffer using BioPython code and that save routine does not close the stream.
Searching for this error and the word alphafold didn't produce anything.
I am suspicious that this is an error produced by Google Colab. Maybe Google Colab is shutting down your virtual machine and it results in this error.
It might be a bit helpful to press on that "4 frames" up/down arrow in the middle of the stack trace to reveal the routines that were being called before overwrite_b_factors().
comment:3 by , 3 years ago
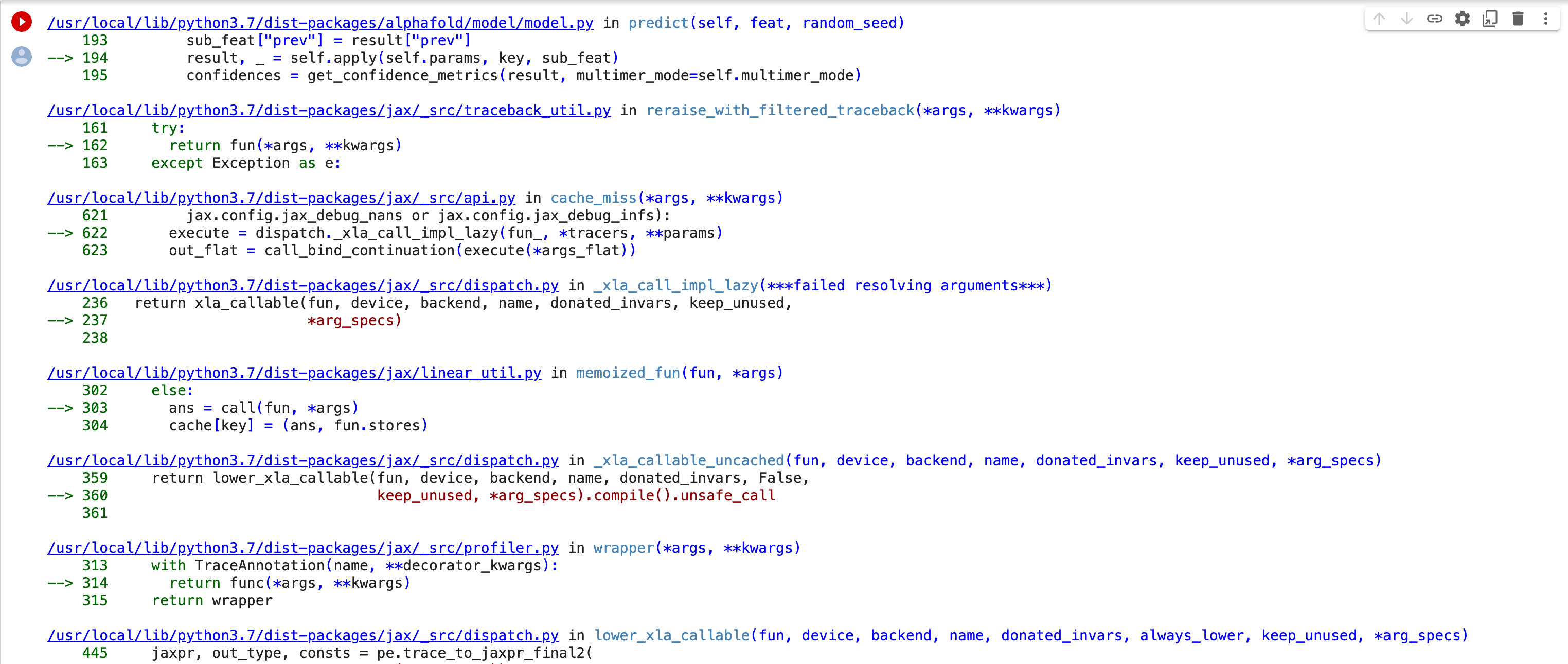
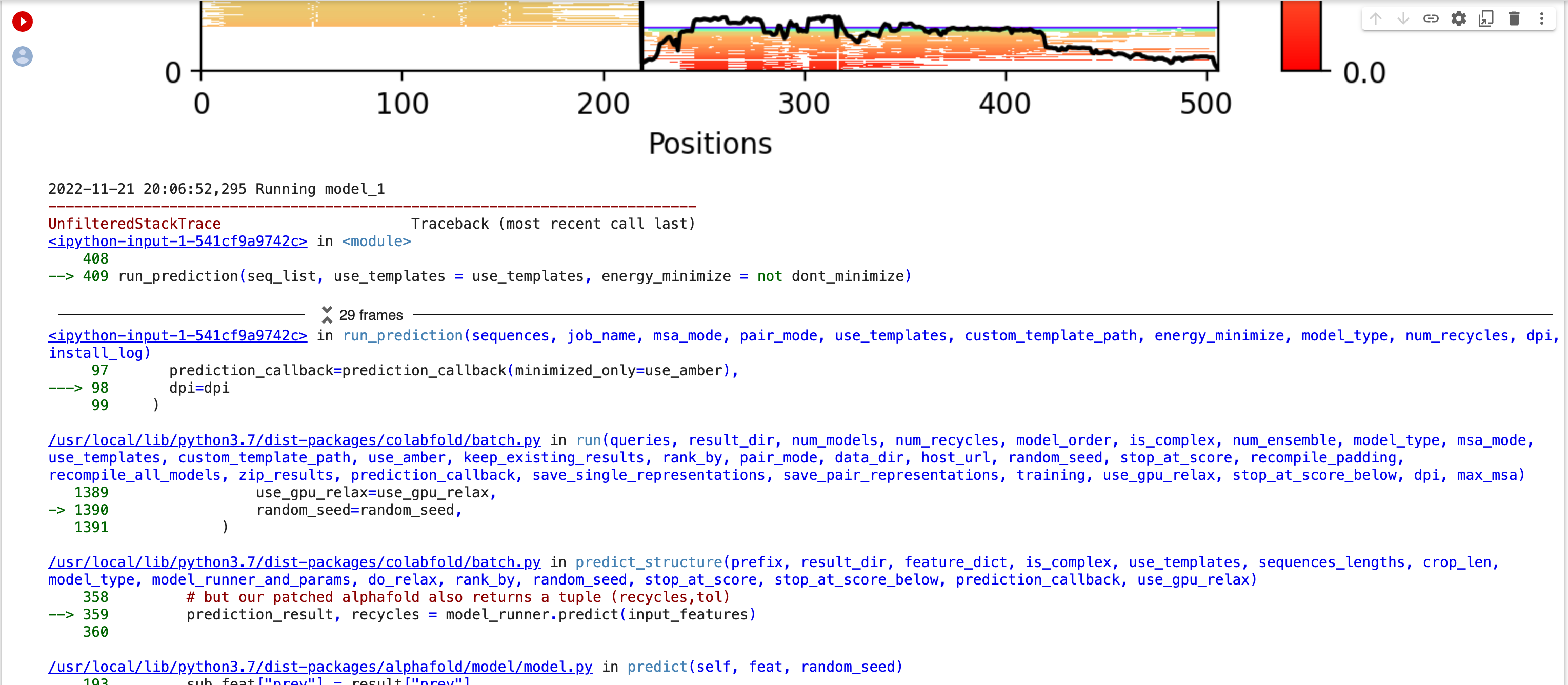
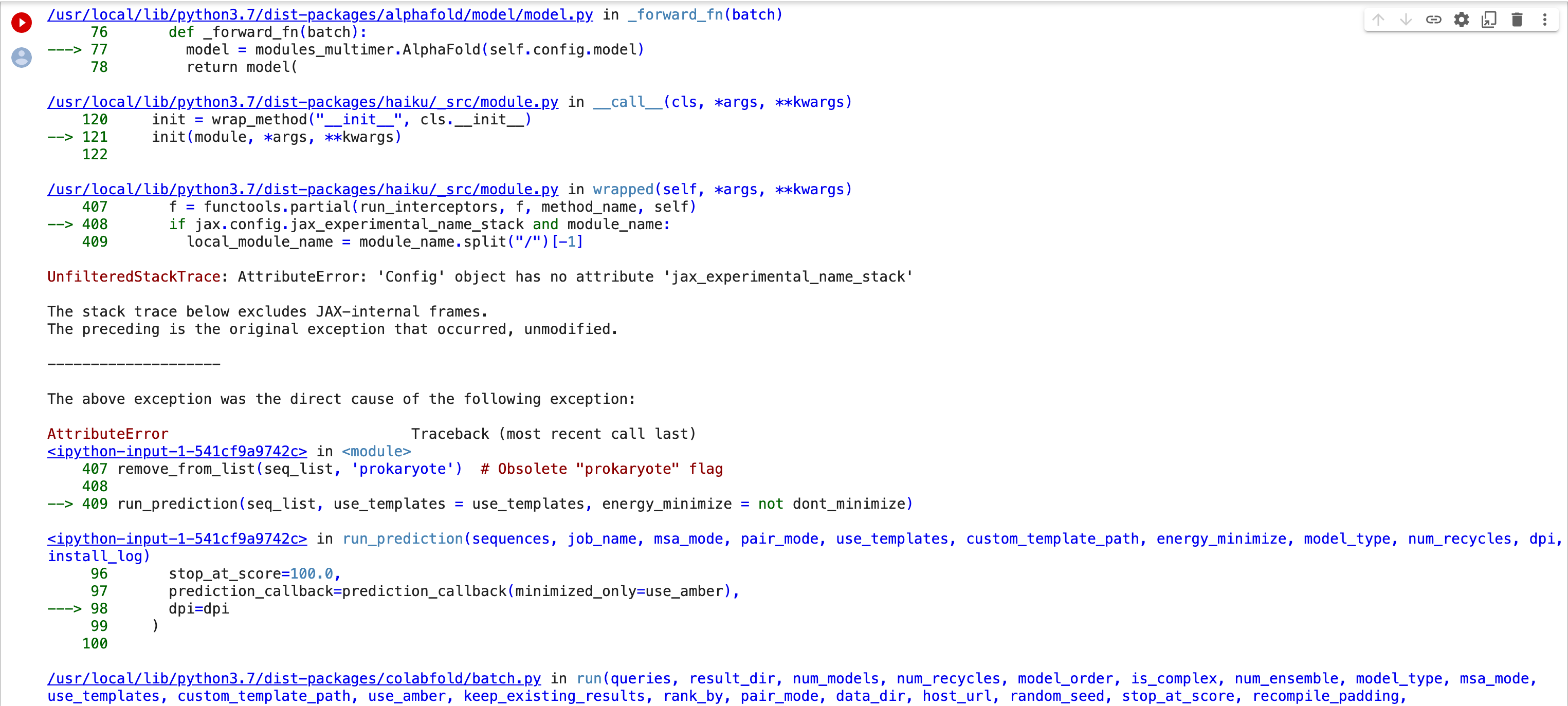
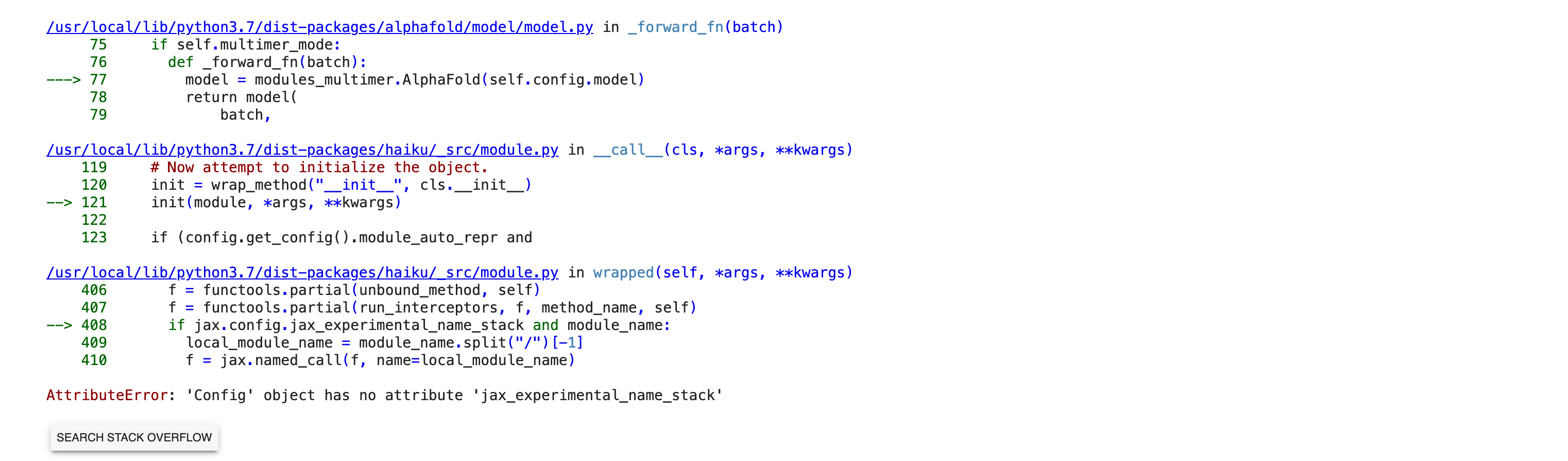
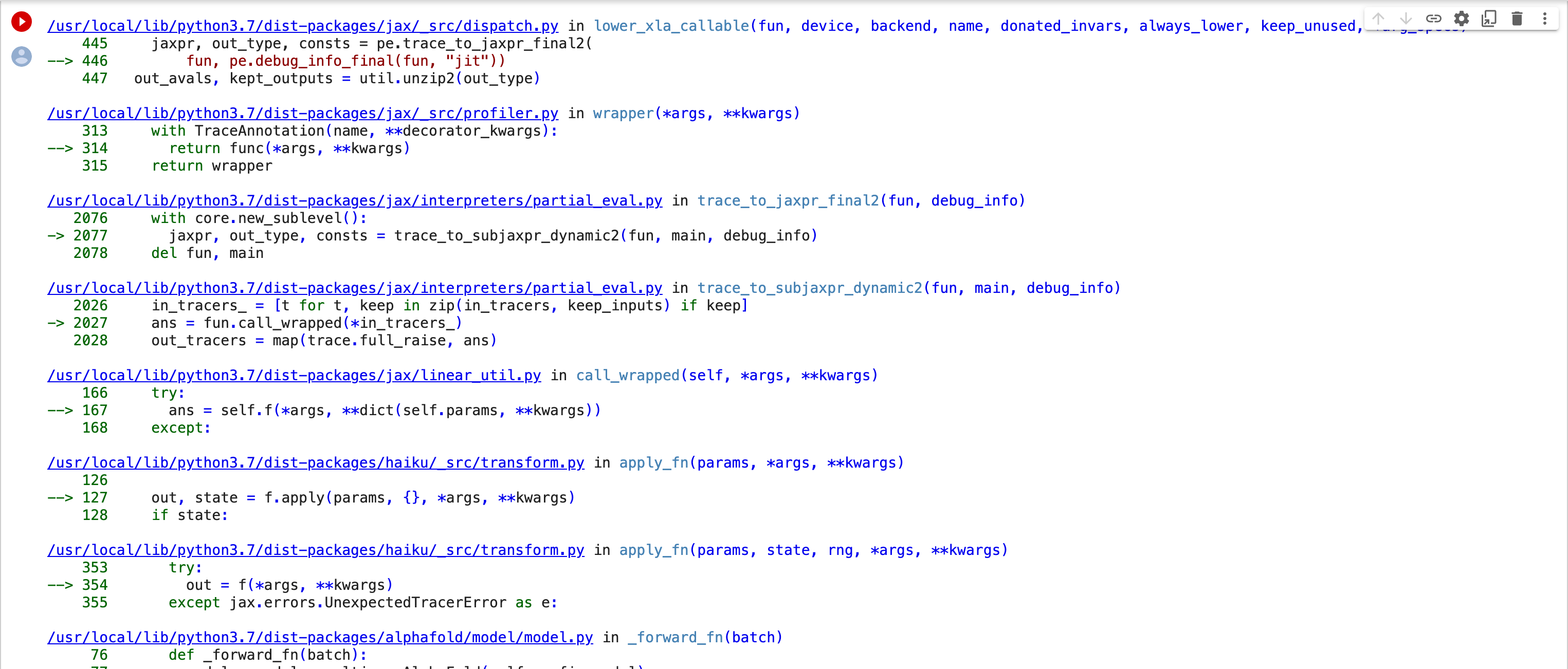
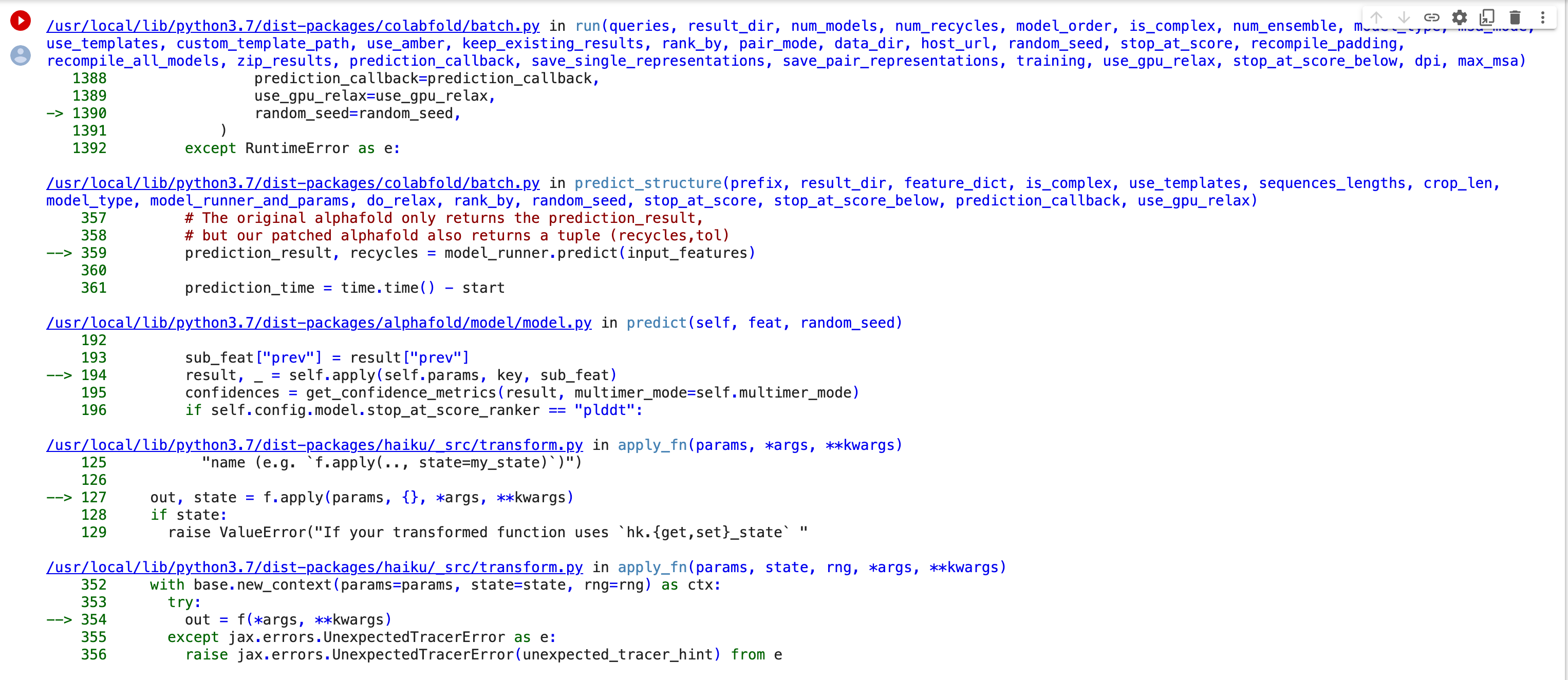
Thank you for looking into this. I re-started a job this morning and got what appears to be a different error. I took screenshots of the "29 frames" in the middle of the stack trace and attached them. The original error had "4 frames". I hope this gets us closer to figuring out the problem. Bernard [image: Screen Shot 2022-11-21 at 1.09.12 PM.png][image: Screen Shot 2022-11-21 at 1.09.29 PM.png][image: Screen Shot 2022-11-21 at 1.09.43 PM.png][image: Screen Shot 2022-11-21 at 1.10.04 PM.png][image: Screen Shot 2022-11-21 at 1.10.18 PM.png][image: Screen Shot 2022-11-21 at 1.10.33 PM.png] On Mon, Nov 21, 2022 at 11:23 AM ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu> wrote:
follow-up: 3 comment:4 by , 3 years ago
This error is apparently caused by an update to a library JAX that AlphaFold uses
https://github.com/deepmind/dm-haiku/issues/565
I also get this error when running AlphaFold in ChimeraX. I will see if ColabFold, which is the AlphaFold package ChimeraX uses has fixed it.
comment:5 by , 3 years ago
The ColabFold prediction web server
https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb
is also failing with the same error. And my test with the latest ColabFold failed with the same error. So apparently they have not yet fixed the problem from the updated jax. So I will try to make a fix forcing ChimeraX ColabFold to use an older JAX.
comment:7 by , 3 years ago
| Resolution: | nonchimerax |
|---|---|
| Status: | closed → reopened |
Fixed.
The release of jax 0.3.25 broke ColabFold. Unfortunately ColabFold asks for jax version >= 0.3.8 and < 4 so whenever jax updates to 0.3.x ColabFold automatically uses it. This has caused ColabFold to break multiple times in the past. So to fix it this time I pinned the jax and jaxlib versions to 0.3.24.
comment:8 by , 3 years ago
| Resolution: | → fixed |
|---|---|
| Status: | reopened → closed |
Added by email2trac