The following bug report has been submitted:
Platform: macOS-10.16-x86_64-i386-64bit
ChimeraX Version: 1.3 (2021-12-08 23:08:33 UTC)
Description
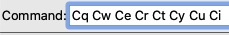
In all text boxes, kerning is weird after some letters. In particular, I notice problems after capital C and D: Cw, Cy, Cz, Cx, and Cv are kerned noticeably too wide. I see this at least in the command line, in this feedback text box and in the model panel. Haven't checked every letter. Thanks!
Log:
Startup Messages
---
note | available bundle cache has not been initialized yet
UCSF ChimeraX version: 1.3 (2021-12-08)
© 2016-2021 Regents of the University of California. All rights reserved.
How to cite UCSF ChimeraX
> open 30xz
Fetching url http://files.rcsb.org/download/30xz.cif failed: HTTP Error 404:
Not Found
> open 3oxz
Summary of feedback from opening 3oxz fetched from pdb
---
notes | Fetching compressed mmCIF 3oxz from
http://files.rcsb.org/download/3oxz.cif
Fetching CCD 0LI from http://ligand-expo.rcsb.org/reports/0/0LI/0LI.cif
3oxz title:
Crystal structure of ABL kinase domain bound with a DFG-out inhibitor AP24534
[more info...]
Chain information for 3oxz #1
---
Chain | Description | UniProt
A | Tyrosine-protein kinase ABL1 | ABL1_MOUSE
Non-standard residues in 3oxz #1
---
0LI —
3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide
(Ponatinib)
> open 3cs9
Summary of feedback from opening 3cs9 fetched from pdb
---
notes | Fetching compressed mmCIF 3cs9 from
http://files.rcsb.org/download/3cs9.cif
Fetching CCD NIL from http://ligand-expo.rcsb.org/reports/N/NIL/NIL.cif
3cs9 title:
Human ABL kinase in complex with nilotinib [more info...]
Chain information for 3cs9 #2
---
Chain | Description | UniProt
A B C D | Proto-oncogene tyrosine-protein kinase ABL1 | ABL1_HUMAN
Non-standard residues in 3cs9 #2
---
NIL — Nilotinib
(4-methyl-N-[3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]-3-[(4-pyridin-3-ylpyrimidin-2-yl)amino]benzamide)
3cs9 mmCIF Assemblies
---
1| author_and_software_defined_assembly
2| author_and_software_defined_assembly
3| author_and_software_defined_assembly
4| author_and_software_defined_assembly
> ui tool show Matchmaker
> matchmaker #!2 to #1
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker 3oxz, chain A (#1) with 3cs9, chain B (#2), sequence alignment
score = 1373.8
RMSD between 242 pruned atom pairs is 0.591 angstroms; (across all 244 pairs:
0.623)
> select #1
2280 atoms, 2249 bonds, 3 pseudobonds, 358 residues, 2 models selected
> ~select #1
Nothing selected
> log metadata #1
Metadata for 3oxz #1
---
Title | Crystal structure of ABL kinase domain bound with a DFG-out inhibitor
AP24534
Citation | Zhou, T., Commodore, L., Huang, W.S., Wang, Y., Thomas, M., Keats,
J., Xu, Q., Rivera, V.M., Shakespeare, W.C., Clackson, T., Dalgarno, D.C.,
Zhu, X. (2011). Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib
(AP24534): Lessons for Overcoming Kinase Inhibitor Resistance. Chem.Biol.Drug
Des., 77, 1-11. PMID: 21118377. DOI: 10.1111/j.1747-0285.2010.01054.x
Non-standard residue | 0LI —
3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide
(Ponatinib)
Gene source | Mus musculus (mouse)
Experimental method | X-ray diffraction
Resolution | 2.2Å
> log chains #1
Chain information for 3oxz #1
---
Chain | Description | UniProt
A | Tyrosine-protein kinase ABL1 | ABL1_MOUSE
> show #!1 target m
> view #1 clip false
> help help:user/tools/modelpanel.html
> hide #!1 target m
> show #!1 target m
> log metadata #1
Metadata for 3oxz #1
---
Title | Crystal structure of ABL kinase domain bound with a DFG-out inhibitor
AP24534
Citation | Zhou, T., Commodore, L., Huang, W.S., Wang, Y., Thomas, M., Keats,
J., Xu, Q., Rivera, V.M., Shakespeare, W.C., Clackson, T., Dalgarno, D.C.,
Zhu, X. (2011). Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib
(AP24534): Lessons for Overcoming Kinase Inhibitor Resistance. Chem.Biol.Drug
Des., 77, 1-11. PMID: 21118377. DOI: 10.1111/j.1747-0285.2010.01054.x
Non-standard residue | 0LI —
3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide
(Ponatinib)
Gene source | Mus musculus (mouse)
Experimental method | X-ray diffraction
Resolution | 2.2Å
> log chains #1
Chain information for 3oxz #1
---
Chain | Description | UniProt
A | Tyrosine-protein kinase ABL1 | ABL1_MOUSE
> log metadata #1
Metadata for 3oxz #1
---
Title | Crystal structure of ABL kinase domain bound with a DFG-out inhibitor
AP24534
Citation | Zhou, T., Commodore, L., Huang, W.S., Wang, Y., Thomas, M., Keats,
J., Xu, Q., Rivera, V.M., Shakespeare, W.C., Clackson, T., Dalgarno, D.C.,
Zhu, X. (2011). Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib
(AP24534): Lessons for Overcoming Kinase Inhibitor Resistance. Chem.Biol.Drug
Des., 77, 1-11. PMID: 21118377. DOI: 10.1111/j.1747-0285.2010.01054.x
Non-standard residue | 0LI —
3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide
(Ponatinib)
Gene source | Mus musculus (mouse)
Experimental method | X-ray diffraction
Resolution | 2.2Å
> log chains #1
Chain information for 3oxz #1
---
Chain | Description | UniProt
A | Tyrosine-protein kinase ABL1 | ABL1_MOUSE
> ui tool show "Selection Inspector"
> log metadata #1
Metadata for 3oxz #1
---
Title | Crystal structure of ABL kinase domain bound with a DFG-out inhibitor
AP24534
Citation | Zhou, T., Commodore, L., Huang, W.S., Wang, Y., Thomas, M., Keats,
J., Xu, Q., Rivera, V.M., Shakespeare, W.C., Clackson, T., Dalgarno, D.C.,
Zhu, X. (2011). Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib
(AP24534): Lessons for Overcoming Kinase Inhibitor Resistance. Chem.Biol.Drug
Des., 77, 1-11. PMID: 21118377. DOI: 10.1111/j.1747-0285.2010.01054.x
Non-standard residue | 0LI —
3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide
(Ponatinib)
Gene source | Mus musculus (mouse)
Experimental method | X-ray diffraction
Resolution | 2.2Å
> log chains #1
Chain information for 3oxz #1
---
Chain | Description | UniProt
A | Tyrosine-protein kinase ABL1 | ABL1_MOUSE
> help help:user
> rename #1 Abl ponatinib
Expected a keyword
> rename #1 "Abl ponatinib"
> rename #2 "Abl nilotinib"
> sym #2
Abl nilotinib mmCIF Assemblies
---
1| author_and_software_defined_assembly| 1 copy of chain A
2| author_and_software_defined_assembly| 1 copy of chain B
3| author_and_software_defined_assembly| 1 copy of chain C
4| author_and_software_defined_assembly| 1 copy of chain D
> log metadata #2
Metadata for Abl nilotinib #2
---
Title | Human ABL kinase in complex with nilotinib
Citations | Weisberg, E., Manley, P.W., Breitenstein, W., Brueggen, J., Cowan-
Jacob, S.W., Ray, A., Huntly, B., Fabbro, D., Fendrich, G., Hall-Meyers, E.,
Kung, A.L., Mestan, J., Daley, G.Q., Callahan, L., Catley, L., Cavazza, C.,
Azam, M., Neuberg, D., Wright, R.D., Gilliland, D.G., Griffin, J.D. (2005).
Characterization of AMN107, a selective inhibitor of native and mutant Bcr-
Abl. Cancer Cell, 7, 129-141. PMID: 15710326. DOI: 10.1016/j.ccr.2005.01.007
Vajpai, N., Strauss, A., Fendrich, G., Cowan-Jacob, S.W., Manley, P.W.,
Grzesiek, S., Jahnke, W. (?). Solution conformations and dynamics of ABL
kinase inhibitor complexes determined by NMR substantiate the different
binding modes of imatinib/nilotinib and dasatinib. To be Published
Non-standard residue | NIL — Nilotinib
(4-methyl-N-[3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]-3-[(4-pyridin-3-ylpyrimidin-2-yl)amino]benzamide)
Gene source | Homo sapiens (human)
Experimental method | X-ray diffraction
Resolution | 2.20Å
> log chains #2
Chain information for Abl nilotinib #2
---
Chain | Description | UniProt
A B C D | Proto-oncogene tyrosine-protein kinase ABL1 | ABL1_HUMAN
> rename #2 "Abl nilotinib 3cs9"
> rename #1 "Abl ponatinib 3oxz"
> open 2gqg
Summary of feedback from opening 2gqg fetched from pdb
---
warning | Atom P has no neighbors to form bonds with according to residue
template for PTR /A:393
notes | Fetching compressed mmCIF 2gqg from
http://files.rcsb.org/download/2gqg.cif
Fetching CCD 1N1 from http://ligand-expo.rcsb.org/reports/1/1N1/1N1.cif
Fetching CCD GOL from http://ligand-expo.rcsb.org/reports/G/GOL/GOL.cif
Fetching CCD PTR from http://ligand-expo.rcsb.org/reports/P/PTR/PTR.cif
2gqg title:
X-ray Crystal Structure of Dasatinib (BMS-354825) Bound to Activated ABL
Kinase Domain [more info...]
Chain information for 2gqg #3
---
Chain | Description | UniProt
A B | Proto-oncogene tyrosine-protein kinase ABL1 | ABL1_HUMAN
Non-standard residues in 2gqg #3
---
1N1 —
N-(2-chloro-6-methylphenyl)-2-({6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-methylpyrimidin-4-yl}amino)-1,3-thiazole-5-carboxamide
(Dasatinib)
GOL — glycerol (glycerin; propane-1,2,3-triol)
2gqg mmCIF Assemblies
---
1| author_defined_assembly
2| author_defined_assembly
> rename #3 "Abl dasatinib 2gqg"
> open 1iep
Summary of feedback from opening 1iep fetched from pdb
---
notes | Fetching compressed mmCIF 1iep from
http://files.rcsb.org/download/1iep.cif
Fetching CCD CL from http://ligand-expo.rcsb.org/reports/C/CL/CL.cif
Fetching CCD STI from http://ligand-expo.rcsb.org/reports/S/STI/STI.cif
1iep title:
Crystal structure of the C-abl kinase domain In complex with sti-571. [more
info...]
Chain information for 1iep #4
---
Chain | Description | UniProt
A B | proto-oncogene tyrosine-protein kinase abl | ABL1_MOUSE
Non-standard residues in 1iep #4
---
CL — chloride ion
STI — 4-(4-methyl-piperazin-1-ylmethyl)-N-[4-methyl-3-(4-pyridin-3-yl-
pyrimidin-2-ylamino)-phenyl]-benzamide (sti-571;imatinib)
1iep mmCIF Assemblies
---
1| author_defined_assembly
2| author_defined_assembly
> rename #4 "Abl imatinib 1iep"
> open 3ue4
Summary of feedback from opening 3ue4 fetched from pdb
---
warning | Unable to fetch template for 'DB8': might have incorrect bonds
note | Fetching compressed mmCIF 3ue4 from
http://files.rcsb.org/download/3ue4.cif
3ue4 title:
Structural and spectroscopic analysis of the kinase inhibitor bosutinib
binding to the Abl tyrosine kinase domain [more info...]
Chain information for 3ue4 #5
---
Chain | Description | UniProt
A B | Tyrosine-protein kinase ABL1 | ABL1_HUMAN
Non-standard residues in 3ue4 #5
---
DB8 —
4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile
(Bosutinib)
3ue4 mmCIF Assemblies
---
1| author_and_software_defined_assembly
2| author_and_software_defined_assembly
> rename #5 "Abl bosutinib 3ue4"
> open 1opl
Summary of feedback from opening 1opl fetched from pdb
---
notes | Fetching compressed mmCIF 1opl from
http://files.rcsb.org/download/1opl.cif
Fetching CCD MYR from http://ligand-expo.rcsb.org/reports/M/MYR/MYR.cif
Fetching CCD P16 from http://ligand-expo.rcsb.org/reports/P/P16/P16.cif
1opl title:
Structural basis for the auto-inhibition of c-Abl tyrosine kinase [more
info...]
Chain information for 1opl #6
---
Chain | Description | UniProt
A B | proto-oncogene tyrosine-protein kinase | ABL1_HUMAN
Non-standard residues in 1opl #6
---
MYR — myristic acid
P16 —
6-(2,6-dichlorophenyl)-2-{[3-(hydroxymethyl)phenyl]amino}-8-methylpyrido[2,3-D]pyrimidin-7(8H)-one
(PD166326)
1opl mmCIF Assemblies
---
1| author_defined_assembly
2| author_and_software_defined_assembly
3| software_defined_assembly
> rename #6 "Abl myristic acid 1opl"
> open 5mo4
Summary of feedback from opening 5mo4 fetched from pdb
---
notes | Fetching compressed mmCIF 5mo4 from
http://files.rcsb.org/download/5mo4.cif
Fetching CCD AY7 from http://ligand-expo.rcsb.org/reports/A/AY7/AY7.cif
5mo4 title:
ABL1 kinase (T334I_D382N) in complex with asciminib and nilotinib [more
info...]
Chain information for 5mo4 #7
---
Chain | Description | UniProt
A | Tyrosine-protein kinase ABL1 | ABL1_HUMAN
Non-standard residues in 5mo4 #7
---
AY7 — asciminib
NIL — Nilotinib
(4-methyl-N-[3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]-3-[(4-pyridin-3-ylpyrimidin-2-yl)amino]benzamide)
> rename #7 "Abl asciminib nilotonib 5mo4"
> ui tool show Matchmaker
> matchmaker #3-4#!2,5-7 to #1
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker Abl ponatinib 3oxz, chain A (#1) with Abl dasatinib 2gqg, chain B
(#3), sequence alignment score = 1361.8
RMSD between 233 pruned atom pairs is 0.585 angstroms; (across all 256 pairs:
2.409)
Matchmaker Abl ponatinib 3oxz, chain A (#1) with Abl imatinib 1iep, chain A
(#4), sequence alignment score = 1423.3
RMSD between 248 pruned atom pairs is 0.529 angstroms; (across all 255 pairs:
0.929)
Matchmaker Abl ponatinib 3oxz, chain A (#1) with Abl nilotinib 3cs9, chain B
(#2), sequence alignment score = 1373.8
RMSD between 242 pruned atom pairs is 0.591 angstroms; (across all 244 pairs:
0.623)
Matchmaker Abl ponatinib 3oxz, chain A (#1) with Abl bosutinib 3ue4, chain B
(#5), sequence alignment score = 1388.5
RMSD between 238 pruned atom pairs is 0.619 angstroms; (across all 252 pairs:
2.418)
Matchmaker Abl ponatinib 3oxz, chain A (#1) with Abl myristic acid 1opl, chain
A (#6), sequence alignment score = 1404.8
RMSD between 230 pruned atom pairs is 0.619 angstroms; (across all 268 pairs:
4.522)
Matchmaker Abl ponatinib 3oxz, chain A (#1) with Abl asciminib nilotonib 5mo4,
chain A (#7), sequence alignment score = 1398.8
RMSD between 227 pruned atom pairs is 0.742 angstroms; (across all 260 pairs:
3.986)
> open 5u8l
Summary of feedback from opening 5u8l fetched from pdb
---
notes | Fetching compressed mmCIF 5u8l from
http://files.rcsb.org/download/5u8l.cif
Fetching CCD O44 from http://ligand-expo.rcsb.org/reports/O/O44/O44.cif
Fetching CCD SO4 from http://ligand-expo.rcsb.org/reports/S/SO4/SO4.cif
5u8l title:
Crystal structure of EGFR kinase domain in complex with a sulfonyl fluoride
probe XO44 [more info...]
Chain information for 5u8l #8
---
Chain | Description | UniProt
A | Epidermal growth factor receptor | EGFR_HUMAN
Non-standard residues in 5u8l #8
---
GOL — glycerol (glycerin; propane-1,2,3-triol)
O44 —
4-[(4-{4-[(3-cyclopropyl-1H-pyrazol-5-yl)amino]-6-[(prop-2-yn-1-yl)carbamoyl]pyrimidin-2-yl}piperazin-1-yl)methyl]benzene-1-sulfonyl
fluoride
SO4 — sulfate ion
> rename #8 "EGFR XO44 5u8l"
> open 1fin
Summary of feedback from opening 1fin fetched from pdb
---
notes | Fetching compressed mmCIF 1fin from
http://files.rcsb.org/download/1fin.cif
Fetching CCD ATP from http://ligand-expo.rcsb.org/reports/A/ATP/ATP.cif
1fin title:
Cyclin A-cyclin-dependent kinase 2 complex [more info...]
Chain information for 1fin #9
---
Chain | Description | UniProt
A C | cyclin-dependent kinase 2 | CDK2_HUMAN
B D | cyclin A | CCNA2_HUMAN
Non-standard residues in 1fin #9
---
ATP — adenosine-5'-triphosphate
1fin mmCIF Assemblies
---
1| author_and_software_defined_assembly
2| author_and_software_defined_assembly
> rename #9 "CDK2 CycA ATP active 1fin"
OpenGL version: 4.1 INTEL-18.5.8
OpenGL renderer: Intel(R) Iris(TM) Plus Graphics 640
OpenGL vendor: Intel Inc.Hardware:
Hardware Overview:
Model Name: MacBook Pro
Model Identifier: MacBookPro14,1
Processor Name: Dual-Core Intel Core i5
Processor Speed: 2.3 GHz
Number of Processors: 1
Total Number of Cores: 2
L2 Cache (per Core): 256 KB
L3 Cache: 4 MB
Hyper-Threading Technology: Enabled
Memory: 8 GB
System Firmware Version: 429.140.8.0.0
OS Loader Version: 540.100.7~23
SMC Version (system): 2.43f11
Software:
System Software Overview:
System Version: macOS 12.3.1 (21E258)
Kernel Version: Darwin 21.4.0
Time since boot: 2:46
Graphics/Displays:
Intel Iris Plus Graphics 640:
Chipset Model: Intel Iris Plus Graphics 640
Type: GPU
Bus: Built-In
VRAM (Dynamic, Max): 1536 MB
Vendor: Intel
Device ID: 0x5926
Revision ID: 0x0006
Metal Family: Supported, Metal GPUFamily macOS 2
Displays:
Color LCD:
Display Type: Built-In Retina LCD
Resolution: 2560 x 1600 Retina
Framebuffer Depth: 24-Bit Color (ARGB8888)
Main Display: Yes
Mirror: Off
Online: Yes
Automatically Adjust Brightness: No
Connection Type: Internal
Locale: (None, 'UTF-8')
PyQt5 5.15.2, Qt 5.15.2
Installed Packages:
alabaster: 0.7.12
appdirs: 1.4.4
appnope: 0.1.2
Babel: 2.9.1
backcall: 0.2.0
blockdiag: 2.0.1
certifi: 2021.5.30
cftime: 1.5.1.1
charset-normalizer: 2.0.9
ChimeraX-AddCharge: 1.2.2
ChimeraX-AddH: 2.1.11
ChimeraX-AlignmentAlgorithms: 2.0
ChimeraX-AlignmentHdrs: 3.2
ChimeraX-AlignmentMatrices: 2.0
ChimeraX-Alignments: 2.2.3
ChimeraX-AlphaFold: 1.0
ChimeraX-AltlocExplorer: 1.0.1
ChimeraX-AmberInfo: 1.0
ChimeraX-Arrays: 1.0
ChimeraX-Atomic: 1.31
ChimeraX-AtomicLibrary: 4.2
ChimeraX-AtomSearch: 2.0
ChimeraX-AtomSearchLibrary: 1.0
ChimeraX-AxesPlanes: 2.0
ChimeraX-BasicActions: 1.1
ChimeraX-BILD: 1.0
ChimeraX-BlastProtein: 2.0
ChimeraX-BondRot: 2.0
ChimeraX-BugReporter: 1.0
ChimeraX-BuildStructure: 2.6.1
ChimeraX-Bumps: 1.0
ChimeraX-BundleBuilder: 1.1
ChimeraX-ButtonPanel: 1.0
ChimeraX-CageBuilder: 1.0
ChimeraX-CellPack: 1.0
ChimeraX-Centroids: 1.2
ChimeraX-ChemGroup: 2.0
ChimeraX-Clashes: 2.2.2
ChimeraX-ColorActions: 1.0
ChimeraX-ColorGlobe: 1.0
ChimeraX-ColorKey: 1.5
ChimeraX-CommandLine: 1.1.5
ChimeraX-ConnectStructure: 2.0
ChimeraX-Contacts: 1.0
ChimeraX-Core: 1.3
ChimeraX-CoreFormats: 1.1
ChimeraX-coulombic: 1.3.2
ChimeraX-Crosslinks: 1.0
ChimeraX-Crystal: 1.0
ChimeraX-CrystalContacts: 1.0
ChimeraX-DataFormats: 1.2.2
ChimeraX-Dicom: 1.0
ChimeraX-DistMonitor: 1.1.5
ChimeraX-DistUI: 1.0
ChimeraX-Dssp: 2.0
ChimeraX-EMDB-SFF: 1.0
ChimeraX-ExperimentalCommands: 1.0
ChimeraX-FileHistory: 1.0
ChimeraX-FunctionKey: 1.0
ChimeraX-Geometry: 1.1
ChimeraX-gltf: 1.0
ChimeraX-Graphics: 1.1
ChimeraX-Hbonds: 2.1.2
ChimeraX-Help: 1.2
ChimeraX-HKCage: 1.3
ChimeraX-IHM: 1.1
ChimeraX-ImageFormats: 1.2
ChimeraX-IMOD: 1.0
ChimeraX-IO: 1.0.1
ChimeraX-ItemsInspection: 1.0
ChimeraX-Label: 1.1
ChimeraX-ListInfo: 1.1.1
ChimeraX-Log: 1.1.4
ChimeraX-LookingGlass: 1.1
ChimeraX-Maestro: 1.8.1
ChimeraX-Map: 1.1
ChimeraX-MapData: 2.0
ChimeraX-MapEraser: 1.0
ChimeraX-MapFilter: 2.0
ChimeraX-MapFit: 2.0
ChimeraX-MapSeries: 2.1
ChimeraX-Markers: 1.0
ChimeraX-Mask: 1.0
ChimeraX-MatchMaker: 2.0.4
ChimeraX-MDcrds: 2.6
ChimeraX-MedicalToolbar: 1.0.1
ChimeraX-Meeting: 1.0
ChimeraX-MLP: 1.1
ChimeraX-mmCIF: 2.4
ChimeraX-MMTF: 2.1
ChimeraX-Modeller: 1.2.6
ChimeraX-ModelPanel: 1.2.1
ChimeraX-ModelSeries: 1.0
ChimeraX-Mol2: 2.0
ChimeraX-Morph: 1.0
ChimeraX-MouseModes: 1.1
ChimeraX-Movie: 1.0
ChimeraX-Neuron: 1.0
ChimeraX-Nucleotides: 2.0.2
ChimeraX-OpenCommand: 1.7
ChimeraX-PDB: 2.6.5
ChimeraX-PDBBio: 1.0
ChimeraX-PDBLibrary: 1.0.2
ChimeraX-PDBMatrices: 1.0
ChimeraX-PickBlobs: 1.0
ChimeraX-Positions: 1.0
ChimeraX-PresetMgr: 1.0.1
ChimeraX-PubChem: 2.1
ChimeraX-ReadPbonds: 1.0.1
ChimeraX-Registration: 1.1
ChimeraX-RemoteControl: 1.0
ChimeraX-ResidueFit: 1.0
ChimeraX-RestServer: 1.1
ChimeraX-RNALayout: 1.0
ChimeraX-RotamerLibMgr: 2.0.1
ChimeraX-RotamerLibsDunbrack: 2.0
ChimeraX-RotamerLibsDynameomics: 2.0
ChimeraX-RotamerLibsRichardson: 2.0
ChimeraX-SaveCommand: 1.5
ChimeraX-SchemeMgr: 1.0
ChimeraX-SDF: 2.0
ChimeraX-Segger: 1.0
ChimeraX-Segment: 1.0
ChimeraX-SelInspector: 1.0
ChimeraX-SeqView: 2.4.6
ChimeraX-Shape: 1.0.1
ChimeraX-Shell: 1.0
ChimeraX-Shortcuts: 1.1
ChimeraX-ShowAttr: 1.0
ChimeraX-ShowSequences: 1.0
ChimeraX-SideView: 1.0
ChimeraX-Smiles: 2.1
ChimeraX-SmoothLines: 1.0
ChimeraX-SpaceNavigator: 1.0
ChimeraX-StdCommands: 1.6.1
ChimeraX-STL: 1.0
ChimeraX-Storm: 1.0
ChimeraX-Struts: 1.0
ChimeraX-Surface: 1.0
ChimeraX-SwapAA: 2.0
ChimeraX-SwapRes: 2.1
ChimeraX-TapeMeasure: 1.0
ChimeraX-Test: 1.0
ChimeraX-Toolbar: 1.1
ChimeraX-ToolshedUtils: 1.2
ChimeraX-Tug: 1.0
ChimeraX-UI: 1.13.7
ChimeraX-uniprot: 2.2
ChimeraX-UnitCell: 1.0
ChimeraX-ViewDockX: 1.0.1
ChimeraX-VIPERdb: 1.0
ChimeraX-Vive: 1.1
ChimeraX-VolumeMenu: 1.0
ChimeraX-VTK: 1.0
ChimeraX-WavefrontOBJ: 1.0
ChimeraX-WebCam: 1.0
ChimeraX-WebServices: 1.0
ChimeraX-Zone: 1.0
colorama: 0.4.4
cxservices: 1.1
cycler: 0.11.0
Cython: 0.29.24
decorator: 5.1.0
docutils: 0.17.1
filelock: 3.0.12
funcparserlib: 0.3.6
grako: 3.16.5
h5py: 3.6.0
html2text: 2020.1.16
idna: 3.3
ihm: 0.21
imagecodecs: 2021.4.28
imagesize: 1.3.0
ipykernel: 5.5.5
ipython: 7.23.1
ipython-genutils: 0.2.0
jedi: 0.18.0
Jinja2: 3.0.1
jupyter-client: 6.1.12
jupyter-core: 4.9.1
kiwisolver: 1.3.2
lxml: 4.6.3
lz4: 3.1.3
MarkupSafe: 2.0.1
matplotlib: 3.4.3
matplotlib-inline: 0.1.3
msgpack: 1.0.2
netCDF4: 1.5.7
networkx: 2.6.3
numexpr: 2.8.0
numpy: 1.21.2
openvr: 1.16.801
packaging: 21.0
ParmEd: 3.2.0
parso: 0.8.3
pexpect: 4.8.0
pickleshare: 0.7.5
Pillow: 8.3.2
pip: 21.2.4
pkginfo: 1.7.1
prompt-toolkit: 3.0.23
psutil: 5.8.0
ptyprocess: 0.7.0
pycollada: 0.7.1
pydicom: 2.1.2
Pygments: 2.10.0
PyOpenGL: 3.1.5
PyOpenGL-accelerate: 3.1.5
pyparsing: 3.0.6
PyQt5-commercial: 5.15.2
PyQt5-sip: 12.8.1
PyQtWebEngine-commercial: 5.15.2
python-dateutil: 2.8.2
pytz: 2021.3
pyzmq: 22.3.0
qtconsole: 5.1.1
QtPy: 1.11.3
RandomWords: 0.3.0
requests: 2.26.0
scipy: 1.7.1
setuptools: 57.5.0
sfftk-rw: 0.7.1
six: 1.16.0
snowballstemmer: 2.2.0
sortedcontainers: 2.4.0
Sphinx: 4.2.0
sphinx-autodoc-typehints: 1.12.0
sphinxcontrib-applehelp: 1.0.2
sphinxcontrib-blockdiag: 2.0.0
sphinxcontrib-devhelp: 1.0.2
sphinxcontrib-htmlhelp: 2.0.0
sphinxcontrib-jsmath: 1.0.1
sphinxcontrib-qthelp: 1.0.3
sphinxcontrib-serializinghtml: 1.1.5
suds-jurko: 0.6
tifffile: 2021.4.8
tinyarray: 1.2.3
tornado: 6.1
traitlets: 5.1.1
urllib3: 1.26.7
wcwidth: 0.2.5
webcolors: 1.11.1
wheel: 0.37.0
wheel-filename: 1.3.0
File attachment: Screen Shot 2022-06-07 at 13.27.04.png
Screen Shot 2022-06-07 at 13.27.04.png
Added by email2trac