Opened 4 years ago
Last modified 4 years ago
#6379 feedback defect
Crash using File→Open on Windows and/or in return statement
| Reported by: | Owned by: | Eric Pettersen | |
|---|---|---|---|
| Priority: | normal | Milestone: | |
| Component: | Platform | Version: | |
| Keywords: | Cc: | Tom Goddard | |
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
The following bug report has been submitted:
Platform: Windows-10-10.0.17763
ChimeraX Version: 1.4.dev202202030703 (2022-02-03 07:03:08 UTC)
Description
Last time you used ChimeraX it crashed.
Please describe steps that led to the crash here.
Windows fatal exception: code 0xe0000002
Thread 0x00001998 (most recent call first):
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\chimerax\open_command\dialog.py", line 135 in show_open_file_dialog
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\chimerax\open_command\dialog.py", line 107 in
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\chimerax\ui\gui.py", line 318 in event_loop
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\ChimeraX_main.py", line 867 in init
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\ChimeraX_main.py", line 1018 in
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\runpy.py", line 87 in _run_code
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\runpy.py", line 197 in _run_module_as_main
Windows fatal exception: code 0xe0000002
Thread 0x00001998 (most recent call first):
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\chimerax\open_command\dialog.py", line 135 in show_open_file_dialog
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\chimerax\open_command\dialog.py", line 107 in
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\chimerax\ui\gui.py", line 318 in event_loop
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\ChimeraX_main.py", line 867 in init
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\ChimeraX_main.py", line 1018 in
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\runpy.py", line 87 in _run_code
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\runpy.py", line 197 in _run_module_as_main
Windows fatal exception: code 0xe0000002
Thread 0x00001998 (most recent call first):
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\chimerax\open_command\dialog.py", line 135 in show_open_file_dialog
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\chimerax\open_command\dialog.py", line 107 in
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\chimerax\ui\gui.py", line 318 in event_loop
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\ChimeraX_main.py", line 867 in init
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\ChimeraX_main.py", line 1018 in
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\runpy.py", line 87 in _run_code
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\runpy.py", line 197 in _run_module_as_main
Windows fatal exception: code 0xe0000002
Thread 0x00001998 (most recent call first):
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\chimerax\open_command\dialog.py", line 135 in show_open_file_dialog
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\chimerax\open_command\dialog.py", line 107 in
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\chimerax\ui\gui.py", line 318 in event_loop
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\ChimeraX_main.py", line 867 in init
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\ChimeraX_main.py", line 1018 in
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\runpy.py", line 87 in _run_code
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\runpy.py", line 197 in _run_module_as_main
Windows fatal exception: code 0xe0000002
Thread 0x00001998 (most recent call first):
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\chimerax\open_command\dialog.py", line 135 in show_open_file_dialog
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\chimerax\open_command\dialog.py", line 107 in
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\chimerax\ui\gui.py", line 318 in event_loop
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\ChimeraX_main.py", line 867 in init
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\site-packages\ChimeraX_main.py", line 1018 in
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\runpy.py", line 87 in _run_code
File "C:\Program Files\ChimeraX 1.4.dev202202030703\bin\lib\runpy.py", line 197 in _run_module_as_main
Windows fatal exception: code 0xe0000002
Thread 0x00001998 (most recent call first):
File "", line 1067 in _handle_fromlist
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: access violation
Windows fatal exception: stack overflow
Windows fatal exception: access violation
Windows fatal exception: access violation
===== Log before crash start =====
Startup Messages
---
warning | Your computer has Intel graphics driver 7262 with a known bug that
causes all Qt user interface panels to be blank. ChimeraX can partially fix
this but may make some panel titlebars and edges black. Hopefully newer Intel
graphics drivers will fix this.
UCSF ChimeraX version: 1.4.dev202202030703 (2022-02-03)
© 2016-2021 Regents of the University of California. All rights reserved.
How to cite UCSF ChimeraX
> open "C:\Users\meetol\Dropbox\CA_projects_Martti\00 G. salaris CAs\Gs BCA
> alone.fasta" format fasta
Summary of feedback from opening C:\Users\meetol\Dropbox\CA_projects_Martti\00
G. salaris CAs\Gs BCA alone.fasta
---
note | Alignment identifier is Gs BCA alone.fasta
Opened 1 sequences from Gs BCA alone.fasta
> open 1ekj format mmcif fromDatabase pdb
Non-standard residues in 1ekj #1
---
ACT — acetate ion
AZI — azide ion
CIT — citric acid
CL — chloride ion
CU — copper (II) ion
EDO — 1,2-ethanediol (ethylene glycol)
ZN — zinc ion
1ekj mmCIF Assemblies
---
1| author_and_software_defined_assembly
2| author_and_software_defined_assembly
> delete #2/D-h
> delete #1/D-h
> delete #1/a-b
> delete #1/D-h
> delete #1:hoh
> select ::name="CL"
1 atom, 1 residue, 1 model selected
> delete #1:cl
> delete #1:cu
> open
> C:/Users/meetol/Downloads/ChimeraX/AlphaFold/prediction_7/best_model.pdb
> format pdb
Chain information for best_model.pdb #2
---
Chain | Description
A | No description available
> ui tool show Matchmaker
> matchmaker #2 to #1
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker 1ekj, chain C (#1) with best_model.pdb, chain A (#2), sequence
alignment score = 249
RMSD between 113 pruned atom pairs is 1.042 angstroms; (across all 200 pairs:
5.616)
> hide target a
> ribbon #1
> ui tool show Matchmaker
> select #2:31-500
3158 atoms, 3192 bonds, 199 residues, 1 model selected
> matchmaker #2 & sel to #1 cutoffDistance 1.3
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 1.3
Matchmaker 1ekj, chain C (#1) with best_model.pdb, chain A (#2), sequence
alignment score = 222.6
RMSD between 85 pruned atom pairs is 0.753 angstroms; (across all 170 pairs:
5.458)
Alignment identifier is 2/A
> select clear
> ui mousemode right zoom
> lighting shadows false
> lighting full
> lighting shadows false
Alignment identifier is 1/C
Destroying pre-existing alignment with identifier 2/A
Alignment identifier is 2/A
> save "C:/Users/meetol/Dropbox/CA_projects_Martti/00 G. salaris CAs/af model
> and pea ca.cxs"
> select #1/C:160
6 atoms, 5 bonds, 1 residue, 1 model selected
> select #1/C:160
6 atoms, 5 bonds, 1 residue, 1 model selected
> select #1/C:162
8 atoms, 7 bonds, 1 residue, 1 model selected
> select #1/C:162
8 atoms, 7 bonds, 1 residue, 1 model selected
> select #1/C:160,162
14 atoms, 12 bonds, 2 residues, 1 model selected
> select #1/C:160,162,164
25 atoms, 22 bonds, 3 residues, 1 model selected
> select #1/C:160,162,164,220
35 atoms, 32 bonds, 4 residues, 1 model selected
> select #1/C:160,162,164,220,223
41 atoms, 37 bonds, 5 residues, 1 model selected
> select #2/A:37
11 atoms, 10 bonds, 1 residue, 1 model selected
> select #2/A:37
11 atoms, 10 bonds, 1 residue, 1 model selected
> select #2/A:37,39
23 atoms, 21 bonds, 2 residues, 1 model selected
> select #2/A:37,39,41
47 atoms, 44 bonds, 3 residues, 1 model selected
> select #2/A:37,39,41,102
64 atoms, 61 bonds, 4 residues, 1 model selected
> select #2/A:37,39,41,102,105
75 atoms, 71 bonds, 5 residues, 1 model selected
> show sel target ab
> select H
1823 atoms, 229 residues, 1 model selected
> hide sel target a
> select #1/C:160,162,164,220
35 atoms, 32 bonds, 4 residues, 1 model selected
> show sel target ab
> style sel stick
Changed 35 atom styles
> cofr #1
> select clear
> select #1/C:160,162,164,220
35 atoms, 32 bonds, 4 residues, 1 model selected
> select #1/C:160,162,164,220,223
41 atoms, 37 bonds, 5 residues, 1 model selected
> show sel target ab
> style sel stick
Changed 41 atom styles
> color sel byhetero
> select clear
> save "C:/Users/meetol/Dropbox/CA_projects_Martti/00 G. salaris CAs/af model
> and pea ca.cxs"
> color #1 forest green
> color #1 byhetero
> toolshed show
> color bfactor
5323 atoms, 449 residues, atom bfactor range 18.7 to 98.5
> select #2
3648 atoms, 3684 bonds, 229 residues, 1 model selected
> color #1 forest green
> color #1 byhetero
> color bfactor sel
3648 atoms, 229 residues, atom bfactor range 50.3 to 98.5
> color #2 gold
> color #2 byhetero
> select :zn
1 atom, 1 residue, 1 model selected
> show sel target ab
> ui tool show "Selection Inspector"
> size sel atomRadius 0.01
Changed 1 atom radii
> size sel atomRadius 0.8
Changed 1 atom radii
> size sel atomRadius 0.9
Changed 1 atom radii
> select clear
> set bgColor white
> graphics silhouettes true
> ui tool show "Selection Inspector"
> select :zn
1 atom, 1 residue, 1 model selected
> size sel atomRadius 0.8
Changed 1 atom radii
> ui tool show "Side View"
> select clear
> graphics depthcue false
Expected a keyword
> lighting depthCue false
> lighting full
> lighting shadows false
Destroying pre-existing alignment with identifier 1/C
Alignment identifier is 1/C
Destroying pre-existing alignment with identifier 2/A
Alignment identifier is 2/A
> select #1/C:329
8 atoms, 7 bonds, 1 residue, 1 model selected
> select #1/C:113-329
1666 atoms, 1711 bonds, 217 residues, 1 model selected
> select #1/C:329
8 atoms, 7 bonds, 1 residue, 1 model selected
> select #1/C:113-329
1666 atoms, 1711 bonds, 217 residues, 1 model selected
> select #1/C:329
8 atoms, 7 bonds, 1 residue, 1 model selected
> select #1/C:113-329
1666 atoms, 1711 bonds, 217 residues, 1 model selected
> select #1/C:259
5 atoms, 4 bonds, 1 residue, 1 model selected
> select #1/C:259
5 atoms, 4 bonds, 1 residue, 1 model selected
> select #1/C:259
5 atoms, 4 bonds, 1 residue, 1 model selected
> select #1/C:259-261
24 atoms, 24 bonds, 3 residues, 1 model selected
> select #2/A:92
20 atoms, 20 bonds, 1 residue, 1 model selected
> select #2/A:92
20 atoms, 20 bonds, 1 residue, 1 model selected
> select #2/A:102
17 atoms, 17 bonds, 1 residue, 1 model selected
> select #2/A:102-106
73 atoms, 73 bonds, 5 residues, 1 model selected
> select #2/A:106
22 atoms, 21 bonds, 1 residue, 1 model selected
> select #2/A:106-112
117 atoms, 116 bonds, 7 residues, 1 model selected
> select #2/A:113
15 atoms, 14 bonds, 1 residue, 1 model selected
> select #2/A:113-120
126 atoms, 125 bonds, 8 residues, 1 model selected
> ui mousemode right translate
> select clear
> ui mousemode right zoom
> save "C:/Users/meetol/Dropbox/CA_projects_Martti/00 G. salaris CAs/af model
> and pea ca v2.cxs"
> save "C:/Users/meetol/Dropbox/CA_projects_Martti/00 G. salaris CAs/GsBCA and
> pea BCA.png" width 1280 height 720 supersample 3
> select #1
1675 atoms, 1717 bonds, 3 pseudobonds, 220 residues, 2 models selected
> ui tool show "Color Actions"
> color #1 sky blue
> save "C:/Users/meetol/Dropbox/CA_projects_Martti/00 G. salaris CAs/GsBCA and
> pea BCA.png" width 1280 height 720 supersample 3
> color #1 heteroatom
Expected a color or one of 'byatom', 'bychain', 'byelement', 'byhetero',
'byidentity', 'bymodel', 'bynucleotide', 'bypolymer', 'fromatoms', or 'random'
or a keyword
> color #1 byhetero
> color :zn red
> select clear
> ui mousemode right translate
> windowsize 900 900
QWindowsWindow::setGeometry: Unable to set geometry 1702x1116+9+38 (frame:
1720x1163+0+0) on QWidgetWindow/"MainWindowClassWindow" on "\\\\.\DISPLAY1".
Resulting geometry: 1702x1055+9+38 (frame: 1720x1102+0+0) margins: 9, 38, 9, 9
minimum size: 470x568 MINMAXINFO maxSize=0,0 maxpos=0,0 mintrack=488,615
maxtrack=0,0)
> ui mousemode right zoom
> ui mousemode right translate
> select #2/A:1
26 atoms, 25 bonds, 1 residue, 1 model selected
> select #2/A:1
26 atoms, 25 bonds, 1 residue, 1 model selected
> select clear
> save "C:/Users/meetol/Dropbox/CA_projects_Martti/00 G. salaris CAs/GsBCA and
> pea BCA.png" width 900 height 839 supersample 3
> select #2/A:2-7,11-26,44-47,69-72,80-91,106-121,130-144,173-195
1564 atoms, 1569 bonds, 96 residues, 1 model selected
> select #2/A:141-142
31 atoms, 30 bonds, 2 residues, 1 model selected
> select #2/A:141-145
88 atoms, 88 bonds, 5 residues, 1 model selected
> select #2/A:141-145
88 atoms, 88 bonds, 5 residues, 1 model selected
> select clear
> select #2/A:2-7,11-26,44-47,69-72,80-91,106-121,130-144,173-195
1564 atoms, 1569 bonds, 96 residues, 1 model selected
> select #2/A:106
22 atoms, 21 bonds, 1 residue, 1 model selected
> select #2/A:106-121
258 atoms, 257 bonds, 16 residues, 1 model selected
> select #2/A:2-7,11-26,44-47,69-72,80-91,106-121,130-144,173-195
1564 atoms, 1569 bonds, 96 residues, 1 model selected
> select #2/A:92
20 atoms, 20 bonds, 1 residue, 1 model selected
> select #2/A:80-92
195 atoms, 196 bonds, 13 residues, 1 model selected
> select #2/A:176
24 atoms, 23 bonds, 1 residue, 1 model selected
> select #2/A:176
24 atoms, 23 bonds, 1 residue, 1 model selected
> select #2/A:2-7,11-26,44-47,69-72,80-91,106-121,130-144,173-195
1564 atoms, 1569 bonds, 96 residues, 1 model selected
> select #2/A:176
24 atoms, 23 bonds, 1 residue, 1 model selected
> select #2/A:176-196
340 atoms, 341 bonds, 21 residues, 1 model selected
> select clear
> ui mousemode right zoom
> save "C:/Users/meetol/Dropbox/CA_projects_Martti/00 G. salaris CAs/af model
> and pea ca v3.cxs"
> windowsize 900 700
> windowsize 700 900
QWindowsWindow::setGeometry: Unable to set geometry 1502x1116+0+29 (frame:
1520x1163-9-9) on QWidgetWindow/"MainWindowClassWindow" on "\\\\.\DISPLAY1".
Resulting geometry: 1502x1055+0+29 (frame: 1520x1102-9-9) margins: 9, 38, 9, 9
minimum size: 470x568 MINMAXINFO maxSize=0,0 maxpos=0,0 mintrack=488,615
maxtrack=0,0)
> windowsize 700 900
QWindowsWindow::setGeometry: Unable to set geometry 1502x1116+0+29 (frame:
1520x1163-9-9) on QWidgetWindow/"MainWindowClassWindow" on "\\\\.\DISPLAY1".
Resulting geometry: 1502x1055+0+29 (frame: 1520x1102-9-9) margins: 9, 38, 9, 9
minimum size: 470x568 MINMAXINFO maxSize=0,0 maxpos=0,0 mintrack=488,615
maxtrack=0,0)
> ui mousemode right translate
> ui mousemode right zoom
> windowsize 650 850
QWindowsWindow::setGeometry: Unable to set geometry 1452x1066+0+29 (frame:
1470x1113-9-9) on QWidgetWindow/"MainWindowClassWindow" on "\\\\.\DISPLAY1".
Resulting geometry: 1452x1055+0+29 (frame: 1470x1102-9-9) margins: 9, 38, 9, 9
minimum size: 470x568 MINMAXINFO maxSize=0,0 maxpos=0,0 mintrack=488,615
maxtrack=0,0)
> ui mousemode right translate
> windowsize 650 800
> save "C:/Users/meetol/Dropbox/CA_projects_Martti/00 G. salaris CAs/GsBCA and
> pea BCA 2.png" width 900 height 839 supersample 3
> windowsize 650 800
> save "C:/Users/meetol/Dropbox/CA_projects_Martti/00 G. salaris CAs/GsBCA and
> pea BCA 2.png" width 650 height 800 supersample 3
> save "C:/Users/meetol/Dropbox/CA_projects_Martti/00 G. salaris CAs/GsBCA and
> pea BCA 2 HR.png" width 900 height 1108 supersample 3
> windowsize 650 800
> ui tool show Matchmaker
> matchmaker #2 to #1 cutoffDistance 1.5 showAlignment true
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 1.5
Matchmaker 1ekj, chain C (#1) with best_model.pdb, chain A (#2), sequence
alignment score = 249
Alignment identifier is 1
Showing conservation header ("seq_conservation" residue attribute) for
alignment 1
Hiding conservation header for alignment 1
Chains used in RMSD evaluation for alignment 1: 1ekj #1/C, best_model.pdb #2/A
Showing rmsd header ("seq_rmsd" residue attribute) for alignment 1
RMSD between 97 pruned atom pairs is 0.858 angstroms; (across all 200 pairs:
5.584)
> windowsize 650 800
> lighting full
> lighting shadows false
> save "C:/Users/meetol/Dropbox/CA_projects_Martti/00 G. salaris CAs/GsBCA and
> pea BCA 2 HR.png" width 1000 height 1231 supersample 3
> ui mousemode right zoom
> ui mousemode right translate
> save "C:/Users/meetol/Dropbox/CA_projects_Martti/00 G. salaris CAs/chimerax
> test.png" width 1118 height 785 supersample 3
> lighting simple
> lighting soft
> lighting full
> lighting shadows false
> lighting shadows true
> lighting shadows false
> lighting shadows true
> lighting flat
> lighting full
> lighting shadows false
> save "C:/Users/meetol/Dropbox/CA_projects_Martti/00 G. salaris CAs/af model
> and pea ca v4.cxs"
> close session
===== Log before crash end =====
I had just closed everything and was preparing to try a command file. The crash happened when I pressed Browse in the file-open dialog. Could be a problem in my Windows too
Log:
Startup Messages
---
warning | Your computer has Intel graphics driver 7262 with a known bug that
causes all Qt user interface panels to be blank. ChimeraX can partially fix
this but may make some panel titlebars and edges black. Hopefully newer Intel
graphics drivers will fix this.
UCSF ChimeraX version: 1.4.dev202202030703 (2022-02-03)
© 2016-2021 Regents of the University of California. All rights reserved.
How to cite UCSF ChimeraX
OpenGL version: 3.3.0 - Build 26.20.100.7262
OpenGL renderer: Intel(R) HD Graphics 520
OpenGL vendor: Intel
Locale: fi_FI.cp1252
Qt version: PyQt5 5.15.2, Qt 5.15.2
Qt platform: windows
Manufacturer: HP
Model: HP EliteBook 840 G3
OS: Microsoft Windows 10 Enterprise (Build 17763)
Memory: 8,464,723,968
MaxProcessMemory: 137,438,953,344
CPU: 4 Intel(R) Core(TM) i5-6200U CPU @ 2.30GHz
OSLanguage: en-US
Installed Packages:
alabaster: 0.7.12
appdirs: 1.4.4
Babel: 2.9.1
backcall: 0.2.0
blockdiag: 3.0.0
certifi: 2021.10.8
cftime: 1.5.2
charset-normalizer: 2.0.11
ChimeraX-AddCharge: 1.2.3
ChimeraX-AddH: 2.1.11
ChimeraX-AlignmentAlgorithms: 2.0
ChimeraX-AlignmentHdrs: 3.2
ChimeraX-AlignmentMatrices: 2.0
ChimeraX-Alignments: 2.2.3
ChimeraX-AlphaFold: 1.0
ChimeraX-AltlocExplorer: 1.0.1
ChimeraX-AmberInfo: 1.0
ChimeraX-Arrays: 1.0
ChimeraX-Atomic: 1.35
ChimeraX-AtomicLibrary: 5.0
ChimeraX-AtomSearch: 2.0
ChimeraX-AtomSearchLibrary: 1.0
ChimeraX-AxesPlanes: 2.1
ChimeraX-BasicActions: 1.1
ChimeraX-BILD: 1.0
ChimeraX-BlastProtein: 2.0
ChimeraX-BondRot: 2.0
ChimeraX-BugReporter: 1.0
ChimeraX-BuildStructure: 2.6.1
ChimeraX-Bumps: 1.0
ChimeraX-BundleBuilder: 1.1
ChimeraX-ButtonPanel: 1.0
ChimeraX-CageBuilder: 1.0
ChimeraX-CellPack: 1.0
ChimeraX-Centroids: 1.2
ChimeraX-ChemGroup: 2.0
ChimeraX-Clashes: 2.2.2
ChimeraX-ColorActions: 1.0
ChimeraX-ColorGlobe: 1.0
ChimeraX-ColorKey: 1.5.1
ChimeraX-CommandLine: 1.2
ChimeraX-ConnectStructure: 2.0
ChimeraX-Contacts: 1.0
ChimeraX-Core: 1.4.dev202202030703
ChimeraX-CoreFormats: 1.1
ChimeraX-coulombic: 1.3.2
ChimeraX-Crosslinks: 1.0
ChimeraX-Crystal: 1.0
ChimeraX-CrystalContacts: 1.0
ChimeraX-DataFormats: 1.2.2
ChimeraX-Dicom: 1.0
ChimeraX-DistMonitor: 1.1.5
ChimeraX-Dssp: 2.0
ChimeraX-EMDB-SFF: 1.0
ChimeraX-ExperimentalCommands: 1.0
ChimeraX-FileHistory: 1.0
ChimeraX-FunctionKey: 1.0
ChimeraX-Geometry: 1.1
ChimeraX-gltf: 1.0
ChimeraX-Graphics: 1.1
ChimeraX-Hbonds: 2.1.2
ChimeraX-Help: 1.2
ChimeraX-HKCage: 1.3
ChimeraX-IHM: 1.1
ChimeraX-ImageFormats: 1.2
ChimeraX-IMOD: 1.0
ChimeraX-IO: 1.0.1
ChimeraX-ItemsInspection: 1.0
ChimeraX-Label: 1.1
ChimeraX-ListInfo: 1.1.1
ChimeraX-Log: 1.1.5
ChimeraX-LookingGlass: 1.1
ChimeraX-Maestro: 1.8.1
ChimeraX-Map: 1.1
ChimeraX-MapData: 2.0
ChimeraX-MapEraser: 1.0
ChimeraX-MapFilter: 2.0
ChimeraX-MapFit: 2.0
ChimeraX-MapSeries: 2.1
ChimeraX-Markers: 1.0
ChimeraX-Mask: 1.0
ChimeraX-MatchMaker: 2.0.6
ChimeraX-MDcrds: 2.6
ChimeraX-MedicalToolbar: 1.0.1
ChimeraX-Meeting: 1.0
ChimeraX-MLP: 1.1
ChimeraX-mmCIF: 2.6
ChimeraX-MMTF: 2.1
ChimeraX-Modeller: 1.5.1
ChimeraX-ModelPanel: 1.3.1
ChimeraX-ModelSeries: 1.0
ChimeraX-Mol2: 2.0
ChimeraX-Morph: 1.0
ChimeraX-MouseModes: 1.1
ChimeraX-Movie: 1.0
ChimeraX-Neuron: 1.0
ChimeraX-Nucleotides: 2.0.2
ChimeraX-OpenCommand: 1.8
ChimeraX-PDB: 2.6.5
ChimeraX-PDBBio: 1.0
ChimeraX-PDBLibrary: 1.0.2
ChimeraX-PDBMatrices: 1.0
ChimeraX-PickBlobs: 1.0
ChimeraX-Positions: 1.0
ChimeraX-PresetMgr: 1.1
ChimeraX-PubChem: 2.1
ChimeraX-ReadPbonds: 1.0.1
ChimeraX-Registration: 1.1
ChimeraX-RemoteControl: 1.0
ChimeraX-ResidueFit: 1.0
ChimeraX-RestServer: 1.1
ChimeraX-RNALayout: 1.0
ChimeraX-RotamerLibMgr: 2.0.1
ChimeraX-RotamerLibsDunbrack: 2.0
ChimeraX-RotamerLibsDynameomics: 2.0
ChimeraX-RotamerLibsRichardson: 2.0
ChimeraX-SaveCommand: 1.5
ChimeraX-SchemeMgr: 1.0
ChimeraX-SDF: 2.0
ChimeraX-Segger: 1.0
ChimeraX-Segment: 1.0
ChimeraX-SelInspector: 1.0
ChimeraX-SeqView: 2.4.6
ChimeraX-Shape: 1.0.1
ChimeraX-Shell: 1.0
ChimeraX-Shortcuts: 1.1
ChimeraX-ShowAttr: 1.0
ChimeraX-ShowSequences: 1.0
ChimeraX-SideView: 1.0
ChimeraX-Smiles: 2.1
ChimeraX-SmoothLines: 1.0
ChimeraX-SpaceNavigator: 1.0
ChimeraX-StdCommands: 1.7.5
ChimeraX-STL: 1.0
ChimeraX-Storm: 1.0
ChimeraX-StructMeasure: 1.0.1
ChimeraX-Struts: 1.0.1
ChimeraX-Surface: 1.0
ChimeraX-SwapAA: 2.0
ChimeraX-SwapRes: 2.1.1
ChimeraX-TapeMeasure: 1.0
ChimeraX-Test: 1.0
ChimeraX-Toolbar: 1.1
ChimeraX-ToolshedUtils: 1.2.1
ChimeraX-Tug: 1.0
ChimeraX-UI: 1.16
ChimeraX-uniprot: 2.2
ChimeraX-UnitCell: 1.0
ChimeraX-ViewDockX: 1.1
ChimeraX-VIPERdb: 1.0
ChimeraX-Vive: 1.1
ChimeraX-VolumeMenu: 1.0
ChimeraX-VTK: 1.0
ChimeraX-WavefrontOBJ: 1.0
ChimeraX-WebCam: 1.0
ChimeraX-WebServices: 1.0
ChimeraX-Zone: 1.0
colorama: 0.4.4
comtypes: 1.1.10
cxservices: 1.1
cycler: 0.11.0
Cython: 0.29.26
debugpy: 1.5.1
decorator: 5.1.1
docutils: 0.17.1
entrypoints: 0.4
filelock: 3.4.2
fonttools: 4.29.1
funcparserlib: 1.0.0a0
grako: 3.16.5
h5py: 3.6.0
html2text: 2020.1.16
idna: 3.3
ihm: 0.26
imagecodecs: 2021.11.20
imagesize: 1.3.0
ipykernel: 6.6.1
ipython: 7.31.1
ipython-genutils: 0.2.0
jedi: 0.18.1
Jinja2: 3.0.3
jupyter-client: 7.1.0
jupyter-core: 4.9.1
kiwisolver: 1.3.2
line-profiler: 3.4.0
lxml: 4.7.1
lz4: 3.1.10
MarkupSafe: 2.0.1
matplotlib: 3.5.1
matplotlib-inline: 0.1.3
msgpack: 1.0.3
nest-asyncio: 1.5.4
netCDF4: 1.5.8
networkx: 2.6.3
numexpr: 2.8.1
numpy: 1.22.1
openvr: 1.16.802
packaging: 21.3
ParmEd: 3.4.3
parso: 0.8.3
pickleshare: 0.7.5
Pillow: 9.0.0
pip: 21.3.1
pkginfo: 1.8.2
prompt-toolkit: 3.0.26
psutil: 5.9.0
pycollada: 0.7.2
pydicom: 2.2.2
Pygments: 2.11.2
PyOpenGL: 3.1.5
PyOpenGL-accelerate: 3.1.5
pyparsing: 3.0.7
PyQt5-commercial: 5.15.2
PyQt5-sip: 12.8.1
PyQtWebEngine-commercial: 5.15.2
python-dateutil: 2.8.2
pytz: 2021.3
pywin32: 303
pyzmq: 22.3.0
qtconsole: 5.2.2
QtPy: 2.0.1
RandomWords: 0.3.0
requests: 2.27.1
scipy: 1.7.3
setuptools: 59.8.0
sfftk-rw: 0.7.1
six: 1.16.0
snowballstemmer: 2.2.0
sortedcontainers: 2.4.0
Sphinx: 4.3.2
sphinx-autodoc-typehints: 1.15.2
sphinxcontrib-applehelp: 1.0.2
sphinxcontrib-blockdiag: 3.0.0
sphinxcontrib-devhelp: 1.0.2
sphinxcontrib-htmlhelp: 2.0.0
sphinxcontrib-jsmath: 1.0.1
sphinxcontrib-qthelp: 1.0.3
sphinxcontrib-serializinghtml: 1.1.5
suds-community: 1.0.0
tables: 3.7.0
tifffile: 2021.11.2
tinyarray: 1.2.4
tornado: 6.1
traitlets: 5.1.1
urllib3: 1.26.8
wcwidth: 0.2.5
webcolors: 1.11.1
wheel: 0.37.1
wheel-filename: 1.3.0
WMI: 1.5.1
Attachments (3)
Change History (13)
comment:1 by , 4 years ago
| Cc: | added |
|---|---|
| Component: | Unassigned → Platform |
| Owner: | set to |
| Platform: | → all |
| Project: | → ChimeraX |
| Status: | new → accepted |
| Summary: | ChimeraX bug report submission → Crash using File→Open on Windows and/or in return statement |
comment:2 by , 4 years ago
| Status: | accepted → feedback |
|---|
follow-up: 3 comment:3 by , 4 years ago
Hi Eric, I think I was trying to change folders using the Windows frequently accessed folders quick link, and it was for opening a file. -Martti ti 15. maalisk. 2022 klo 18.59 ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu> kirjoitti:
follow-up: 4 comment:4 by , 4 years ago
comment:5 by , 4 years ago
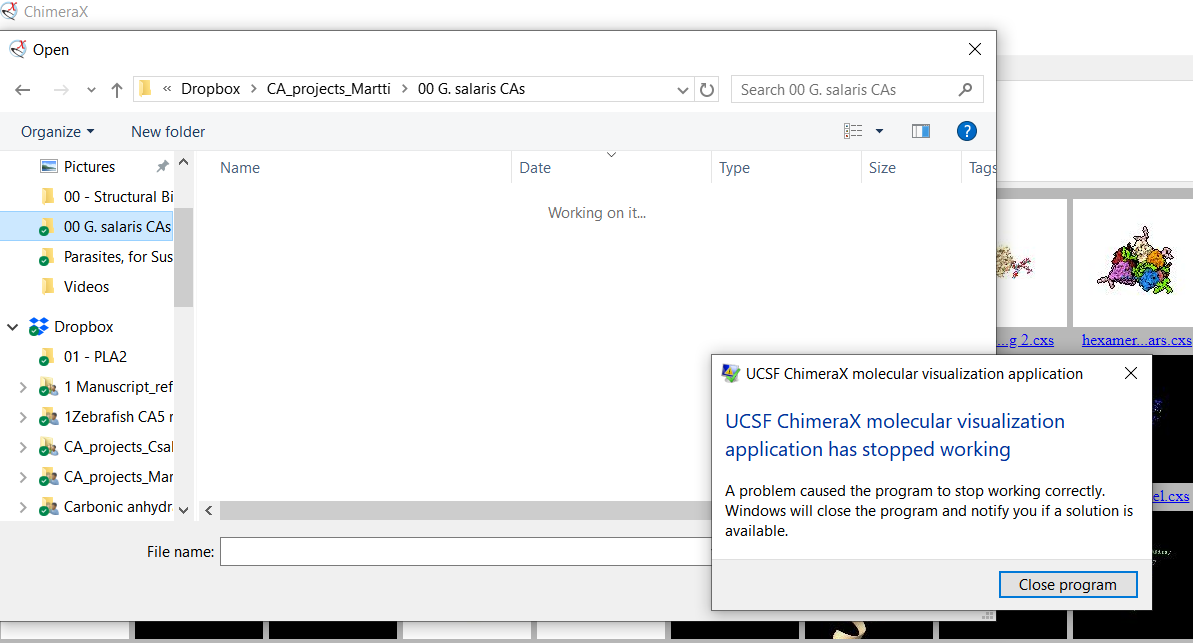
I was able to reproduce the crash on a first try, and this time it was the
first thing I did in a ChimeraX session (same build of 1.14 as in the bug
report). Here's the pic. What happened was that I just clicked on the
folder quick link and after maybe 5 seconds of trying to load the folder
contents ChimeraX crashed. The link is not a pinned one, just one that
Windows has created automatically due to frequent file access recently. The
folder contains only 90 files and no subfolders, and the Dropbox folder you
see in the address bar is right below C:\Users\[my_username]\. As a
non-expert guess, maybe these quick links contain the pathname of ~
implicitly in a way that is not passed on to ChimeraX?
[image: image.png]
I have a new laptop waiting to be taken into use, with a newer version of
Windows 10 which might remove the problem. I also get occasional crashes of
the File Explorer outside of any application in this computer, not related
to these quick links for frequently used folders, but some other random
events that I haven't really tried to follow up. What happens is that all
File Explorer (Windows Explorer?) windows are closed but Windows keeps
running, and I can reopen whichever folders I need and go on working
without needing to reboot.
BTW while I'm at it, I could report a bug (or feature) in Chimera (not X)
new installations which replace a previous version. When I install a new
version of Chimera, opening a file (of a type registered to Chimera) stops
working by double-clicking from a Windows folder, or by choosing Chimera
from the "open with" dialog. What is more, trying to teach Windows to use
the new version of Chimera ("use this application for opening files of this
type") didn't work either. This is because the registry entry
Computer\HKEY_USERS\S-1-5-21-1004336348-152049171-1801674531-111748_Classes\Applications\chimera.exe\shell\open\command
is left behind with a pointer to the previous installation folder of
Chimera. In my case the value was "C:\Program Files\Chimera
1.15\bin\chimera.exe" "%1" until a few minutes ago, pointing to the
previous location which is a non-existent folder now. After editing it (in
regedit) into "C:\Program Files\Chimera 1.16\bin\chimera.exe" "%1", just
changing 1.15 to 1.16 in the pathname, Chimera starts launching as
expected. This annoyance has been around at least since Chimera 1.12, and
I've just resigned to opening files only from inside of Chimera until I get
the inspiration to dig into the registry (as I did now).
Cheers,
-Martti
pe 18. maalisk. 2022 klo 2.36 Eric Pettersen (pett@cgl.ucsf.edu) kirjoitti:
follow-up: 5 comment:6 by , 4 years ago
Hi Martti,
We recently upgraded the version of Python that ChimeraX uses (3.9.6→3.9.11) and are hopeful that certain specific bug fixes that are included with that upgrade may remedy this problem. We are hoping you can try out the latest daily build and see if the Open dialog seems more stable...
--Eric
follow-up: 9 comment:7 by , 4 years ago
Hi Eric, I finally had a chance to try the latest daily version, and now file-open from those quick link folders works, from the same old laptop. Thanks! I was also excited to find the alphafold error plot tool, and happy to see "show all colors" in the color action menu. I can also see that those JSON files are a new thing from AlphaFold, they were not downloaded with a prediction ~3 weeks ago but were there two days ago. Did you ask for them? Cheers, -Martti ke 6. huhtik. 2022 klo 21.30 ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu> kirjoitti:
comment:8 by , 4 years ago
I updated ChimeraX AlphaFold predictions using Colab to use AlphaFold 2.2.0 earlier this week, and I made it use the "monomer_ptm" preset instead of the "monomer" preset since the the latter does not compute PAE and the json files. Those changes were in the ChimeraX Colab script which ChimeraX fetches from GitHub when you make a prediction, so those updates even apply to older ChimeraX daily builds. But the AlphaFold Error Plot button and ability to show those plots requires a daily build from this week. Here's a web page explaining the new error plots and a video demonstrating them if you are interested in more info https://www.rbvi.ucsf.edu/chimerax/data/pae-apr2022/pae.html <https://www.rbvi.ucsf.edu/chimerax/data/pae-apr2022/pae.html> https://youtu.be/oxblwn0_PMM
comment:9 by , 4 years ago
Well, that's encouraging! Have my fingers crossed that it fixes it for everyone -- since we get this reported multiple times per day!
--Eric
Replying to martti.tolvanen@…:
Hi Eric, I finally had a chance to try the latest daily version, and now file-open from those quick link folders works, from the same old laptop. Thanks! I was also excited to find the alphafold error plot tool, and happy to see "show all colors" in the color action menu. I can also see that those JSON files are a new thing from AlphaFold, they were not downloaded with a prediction ~3 weeks ago but were there two days ago. Did you ask for them? Cheers, -Martti ke 6. huhtik. 2022 klo 21.30 ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu> kirjoitti:
follow-up: 7 comment:10 by , 4 years ago
I had the crash happen once again some days ago. I think it was when I changed the folder display from icons into details. But I was too busy to try to repeat that at the time. I'll let you know if I can repeat the precise actions next week. Happy Easter, -Martti ti 12. huhtik. 2022 klo 3.40 ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu> kirjoitti:
Note:
See TracTickets
for help on using tickets.


Hi Martti,
--Eric