Opened 4 years ago
Closed 4 years ago
#6186 closed defect (fixed)
ghosting of left eye on 3D stereo of a selected atom.
| Reported by: | Owned by: | Tom Goddard | |
|---|---|---|---|
| Priority: | normal | Milestone: | |
| Component: | Graphics | Version: | |
| Keywords: | Cc: | ||
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
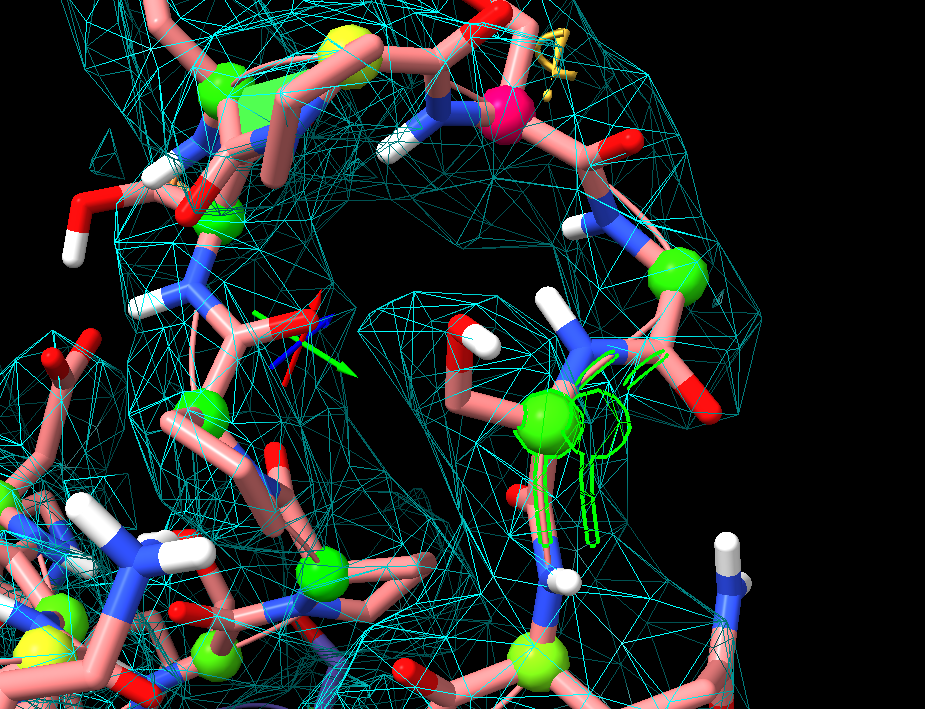
When it is selected, green borders are around the atoms. But in 3D only the right eye has the correct depth. The left eye is left in the 2D plane of the screen. kas [cid:6ab8a262-6de5-4fbe-a424-d0ebd051006f] Kenneth A. Satyshur, M.S., Ph.D. Senior Scientist, College of Ag and Life Sciences: Department of Bacteriology; School of Medicine and Public Health: Departments of Biomolecular Chemistry, Neuroscience, Oncology, and Carbone Cancer Center (Small Molecule Screening Facility) University of Wisconsin-Madison Madison, Wisconsin, 53706 608-215-5207
Attachments (1)
Change History (10)
by , 4 years ago
| Attachment: | ghost-isolde.png added |
|---|
comment:1 by , 4 years ago
| Component: | Unassigned → Graphics |
|---|---|
| Owner: | set to |
| Platform: | → all |
| Project: | → ChimeraX |
| Status: | new → assigned |
comment:2 by , 4 years ago
Hi Ken,
Sorry I have not responded to your stereo bug reports from the last few days. We are trying to renew almost all the ChimeraX funding with proposals due in by the end of the month so things are frenetic. And on top of that I have been preparing a VR demo for the NIH all this week. So it may be next week before I can look at these (and dozens of other) bug reports.
Tom
comment:3 by , 4 years ago
Thanks for the reply. no hurry. Good luck with the demo.
kas
Kenneth A. Satyshur, M.S., Ph.D.
Senior Scientist,
College of Ag and Life Sciences: Department of Bacteriology;
School of Medicine and Public Health:
Departments of Biomolecular Chemistry,
Neuroscience, Oncology, and Carbone Cancer Center
(Small Molecule Screening Facility)
University of Wisconsin-Madison
Madison, Wisconsin, 53706
608-215-5207
________________________________
From: ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu>
Sent: Thursday, February 17, 2022 12:29 PM
Cc: goddard@cgl.ucsf.edu <goddard@cgl.ucsf.edu>; Kenneth Satyshur <kenneth.satyshur@wisc.edu>
Subject: Re: [ChimeraX] #6186: ghosting of left eye on 3D stereo of a selected atom.
#6186: ghosting of left eye on 3D stereo of a selected atom.
-----------------------------------------+-------------------------
Reporter: kenneth.satyshur@… | Owner: Tom Goddard
Type: defect | Status: assigned
Priority: normal | Milestone:
Component: Graphics | Version:
Resolution: | Keywords:
Blocked By: | Blocking:
Notify when closed: | Platform: all
Project: ChimeraX |
-----------------------------------------+-------------------------
Comment (by Tom Goddard):
Hi Ken,
Sorry I have not responded to your stereo bug reports from the last few
days. We are trying to renew almost all the ChimeraX funding with
proposals due in by the end of the month so things are frenetic. And on
top of that I have been preparing a VR demo for the NIH all this week. So
it may be next week before I can look at these (and dozens of other) bug
reports.
Tom
--
Ticket URL: <https://www.rbvi.ucsf.edu/trac/ChimeraX/ticket/6186#comment:2>
ChimeraX <https://www.rbvi.ucsf.edu/chimerax/>
ChimeraX Issue Tracker
follow-up: 3 comment:4 by , 4 years ago
Not sure I understand this bug report. I see two images (left and right eye) for the selection outline, but I don't see two views of the atoms. Maybe that is the problem, the left eye atom rendering is blank except for the selection outline. This may be a manifestation of but #6171, although if that error was encountered I would expect no rendering at all.
This will require testing on our only machine (vive.cgl.ucsf.edu) with a sequential stereo setup and that machine has not been used for that in at least 2 years so may not have drivers and emitter working. So the first challenge will be even testing to reproduce this bug. I am rarely on campus to use that machine due to the pandemic.
comment:5 by , 4 years ago
Thanks Tom and to all the techies at Chimeraland! You have done a great job on chimera and chimeraX. It is a mainstream density fitting for cryoEM, and I have used it as such. But 3D stereo 'the-old-way' using quadro Nvidia cards and Nvidia 3D hardware is not dead yet. I have 6 systems on campus that use the old gamers way of seeing depth. Unfortunately, Nvidia sees a profit in AI, GPU computation, Cryto mining and VR. So what can some old guys do? Just hang onto the legacy systems as long as is practical. I don't see any usefulness to VR is molecular modelling. I don't see Schrodinger rushing to implement VR. Maybe someone who likes the technology should give a demo at the ACA meeting on use of VR to fit CryoEM density. I am going to try to get the Wisconsin Institutes for Discovery (WID) to install ChimeraX and Isolde and demo VR for me. They use it for a virtual tours of tourist attractions.
Being in X-ray crystallography, I use coot to fit density and it has evolved into a powerful 3D refinement package, along with Phenix. I recently used chimera (not chimeraX) to move my models into CryoEM density, Then refined its position, one domain at a time, with Phenix. I then used coot and the sphere-refine system to final fit of the DNA and protein. I works out great and I can do it in 3D. Done in 2 days, led to a successful grant application and a very happy professor.
So if there is anyone that want's to learn the legacy system, let me know. I have a short but bumpy document on the steps to take.
thanks
kas
Kenneth A. Satyshur, M.S., Ph.D.
Senior Scientist,
College of Ag and Life Sciences: Department of Bacteriology;
School of Medicine and Public Health:
Departments of Biomolecular Chemistry,
Neuroscience, Oncology, and Carbone Cancer Center
(Small Molecule Screening Facility)
University of Wisconsin-Madison
Madison, Wisconsin, 53706
608-215-5207
________________________________
From: ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu>
Sent: Saturday, February 19, 2022 2:02 AM
Cc: goddard@cgl.ucsf.edu <goddard@cgl.ucsf.edu>; Kenneth Satyshur <kenneth.satyshur@wisc.edu>
Subject: Re: [ChimeraX] #6186: ghosting of left eye on 3D stereo of a selected atom.
#6186: ghosting of left eye on 3D stereo of a selected atom.
-----------------------------------------+-------------------------
Reporter: kenneth.satyshur@… | Owner: Tom Goddard
Type: defect | Status: assigned
Priority: normal | Milestone:
Component: Graphics | Version:
Resolution: | Keywords:
Blocked By: | Blocking:
Notify when closed: | Platform: all
Project: ChimeraX |
-----------------------------------------+-------------------------
Comment (by Tom Goddard):
Not sure I understand this bug report. I see two images (left and right
eye) for the selection outline, but I don't see two views of the atoms.
Maybe that is the problem, the left eye atom rendering is blank except for
the selection outline. This may be a manifestation of but #6171, although
if that error was encountered I would expect no rendering at all.
This will require testing on our only machine (vive.cgl.ucsf.edu) with a
sequential stereo setup and that machine has not been used for that in at
least 2 years so may not have drivers and emitter working. So the first
challenge will be even testing to reproduce this bug. I am rarely on
campus to use that machine due to the pandemic.
--
Ticket URL: <https://www.rbvi.ucsf.edu/trac/ChimeraX/ticket/6186#comment:4>
ChimeraX <https://www.rbvi.ucsf.edu/chimerax/>
ChimeraX Issue Tracker
follow-up: 5 comment:6 by , 4 years ago
I also see this bug on our vive.cgl.ucsf.edu Quadro P6000 system in stereo.
Selecting chain A of a structure 5n5f, the selection outline is shown correctly in the right eye, but the left eye shows the selection outline for both the left and right eye views, do two non-fused outlines are shown in the left eye. Apparently the outline rendering does not clear the outline drawn for the right eye and simply adds the outline for the left eye when making the left eye image.
comment:7 by , 4 years ago
Side by side stereo mode (camera sbs) correctly shows the selection outlines.
comment:8 by , 4 years ago
Right eye selection outline appears in both left and right eye images. Left eye selection outline only appears in left eye image. Left eye image is rendered first. The problem is the code is rendering the right eye selection outline with glDrawMode(GL_BACK) instead of with GL_BACK_RIGHT. This wrong mode is set when the framebuffer stack is popped. I tried changing the code to restore the correct GL_BACK_RIGHT and that fixed the selection outline. But then switching to mono camera raises an error because switching camera modes did not restore the mode to GL_BACK. There is not a good place in the code to switch the mode back when the camera switches. A few choices would be in the StereoCamera.delete() method, or it could be reset after every frame is drawn in StereoCamera.combine_rendered_camera_views(). The first approach has the problem that that routine does not have the framebuffer available to switch the mode. Could probably get the default framebuffer but this is hacky. Second choice would probably work.
Will need to try the stereo machine to test either of these fixes.
comment:9 by , 4 years ago
| Resolution: | → fixed |
|---|---|
| Status: | assigned → closed |
Fixed.
Made opengl code use correct glDrawMode() in stereo and when switching back to mono.
Added by email2trac