Opened 4 years ago
Last modified 4 years ago
#6106 assigned enhancement
Efficiently select maps by visual inspection
| Reported by: | Owned by: | Tom Goddard | |
|---|---|---|---|
| Priority: | moderate | Milestone: | |
| Component: | Volume Data | Version: | |
| Keywords: | Cc: | guillaume.gaullier@…, vincent.chaptal@… | |
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
Oliver visually inspects up to 50 aligned maps to find some with certain features, e.g. matching an atomic model. He uses the Chimera checkbuttons below the command line that display and hide individual models with Next and Previous buttons to flip through them, and also the row of "skip" checkbuttons which indicate which models to skip when using Next and Previous buttons. Both these rows of buttons are enabled with Chimera Preferences.
I attached a video showing how Oliver uses this.
Oliver wants a similar capability in ChimeraX.
Attachments (2)
Change History (16)
by , 4 years ago
| Attachment: | next_prev.m4v added |
|---|
comment:1 by , 4 years ago
comment:2 by , 4 years ago
Begin forwarded message:
From: Oliver Clarke
Subject: Re: [chimerax-users] Next/previous buttons?
Date: February 7, 2022 at 6:30:22 AM PST
To: Guillaume Gaullier
Cc: Tom Goddard , vincent Chaptal <, ChimeraX Users Help <chimerax-users@…>
To be clear I'm not saying this is the optimal interface - I agree with Tom that rows of checkboxes are cumbersome - I just want the flexibility and rapid switching afforded by this interface (however it is implemented). Capacity to deal with even more maps/models would be great!
Cheers
Oli
comment:3 by , 4 years ago
Begin forwarded message:
From: Guillaume Gaullier
Subject: Re: [chimerax-users] Next/previous buttons?
Date: February 7, 2022 at 5:42:07 AM PST
To: Tom Goddard
Cc: vincent Chaptal, ChimeraX Users Help <chimerax-users@…>, Oliver Clarke
Sorry, I sent my previous answer before I finished reading the discussion. My use case is mostly to compare many maps that are already aligned (or already close enough that I don’t need to align them anyway), and I need to be able to do things like:
- scroll through all maps (vseries does this very well, with the slider or play button if I want to let it cycle through the series),
- keep any number of maps with arbitrary indices displayed, while scrolling through all other maps,
- display any number of maps with arbitrary indices while hiding all the others
1 is easy to do with vseries. 2 and 3 are doable with the model panel checkboxes, but are tedious and clicking-intensive beyond about 10 maps (and typical work easily gets you 50 maps to look at) since you need to uncheck map n and check map n+1 every time you want to show the next map. 3 is doable with a command, so I should study the model selection syntax more closely and stop complaining. But I can’t see how to do 2 with a command.
So everything I need to do is doable with ChimeraX in its current form, but if some of these things required less clicks I would not shy away from looking at more than 50 maps.
I hope this description helps understanding the use case. Curious to see the screen recording from Oli.
Cheers,
Guillaume
comment:4 by , 4 years ago
Oliver, That video was helpful. So I see you trying to visually inspect all your maps with minimal clicking and select a few of them that meet some criterion you are looking for. It looked painful using those checkbuttons. I can only imagine the groove worn in your desk as you move the mouse back and forth to the Next button if you were doing this with 30 maps. But of course I see how it is vastly more painful to use Model Panel checkbuttons. I wonder if we could make something easier and faster, maybe arrow keys to go forward and back and downarrow to hide and skip or up arrow to show and skip. Maybe something needed to unskip, though it is not clear how that would work without the button for every map.
comment:5 by , 4 years ago
Oliver and Guillame. Do I have it right that your use is to visually filter up to 50 maps to find some with selected properties? You have told me way too much about the requirements and too little about the application. I'd never implement something in ChimeraX that way without understanding the application -- it would just work badly if I don't grasp the use cases and use it myself. Those checkbuttons in Chimera are a good example. I see it is the closest method you have, but it looked really painful the huge number of clicks and mouse moves and if I was doing that more than one day I would surely write some code to do much more efficiently. One other comment -- sadly we are busy trying to renew ChimeraX funding, and not even managing to get done projects that do have funding, so the chance of working on this map selection interface soon is not good.
comment:6 by , 4 years ago
Hi Tom,
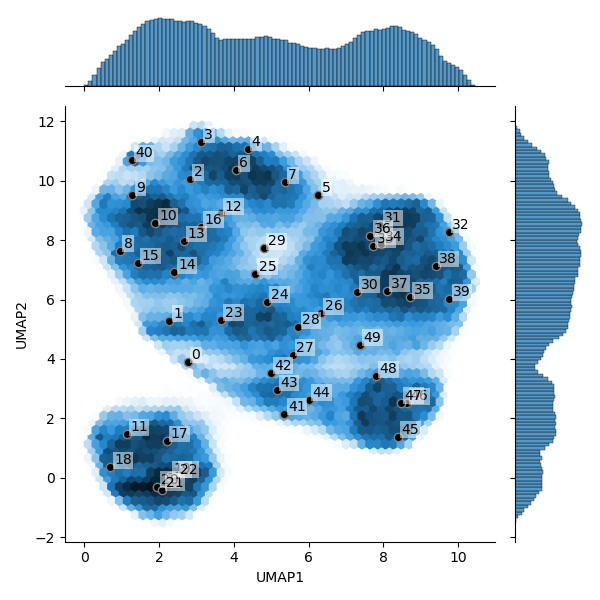
Here is an example of why I need to look at so many maps (and therefore, why it is helpful if switching maps in ChimeraX is as friction-less as possible). I use a program called cryoDRGN (see https://doi.org/10.1038/s41592-020-01049-4 ) that, in brief, determines how heterogeneous a set of particles is, both in terms of composition and conformation. It generates distributions like in the attached picture (darker blue means more particles, white means no particles), in which:
- distinct clusters correspond to populations of particles with different compositions (think complex versus its dissociated components, for example)
- how spread out a given cluster is correlates with how much conformational variability exists in the corresponding ensemble of particles
One thing this program can do is generate maps from this distribution. So, a good way to get a visual overview of the heterogeneity is to randomly take samples from this distribution and generate a map for each one. This is what the numbers on the graph are: each one corresponds to a map I have to look at and compare to the others and/or to an atomic model. The program lets you generate any arbitrary number of maps. When I first used it, I had a distribution small enough that a random sample of 20 maps was enough to get all the info I needed. But when applying this program to larger, more heterogeneous sets of particles, you quickly need to generate many more maps to sample the distribution correctly. The example I attached was decently sampled with 50 maps, but I can easily see how some other cases could require more than that.
I hope this answers your question.
Have a good weekend,
Guillaume
On 8 Feb 2022, at 04:50, ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu<mailto:ChimeraX-bugs-admin@cgl.ucsf.edu>> wrote:
#6106: Efficiently select maps by visual inspection
-----------------------------------+-------------------------
Reporter: olibclarke@… | Owner: Tom Goddard
Type: enhancement | Status: assigned
Priority: moderate | Milestone:
Component: Volume Data | Version:
Resolution: | Keywords:
Blocked By: | Blocking:
Notify when closed: | Platform: all
Project: ChimeraX |
-----------------------------------+-------------------------
Comment (by Tom Goddard):
Oliver and Guillame. Do I have it right that your use is to visually
filter up to 50 maps to find some with selected properties? You have told
me way too much about the requirements and too little about the
application. I'd never implement something in ChimeraX that way without
understanding the application -- it would just work badly if I don't grasp
the use cases and use it myself. Those checkbuttons in Chimera are a good
example. I see it is the closest method you have, but it looked really
painful the huge number of clicks and mouse moves and if I was doing that
more than one day I would surely write some code to do much more
efficiently. One other comment -- sadly we are busy trying to renew
ChimeraX funding, and not even managing to get done projects that do have
funding, so the chance of working on this map selection interface soon is
not good.
--
Ticket URL: <https://www.rbvi.ucsf.edu/trac/ChimeraX/ticket/6106#comment:5>
ChimeraX <https://www.rbvi.ucsf.edu/chimerax/>
ChimeraX Issue Tracker
[cid:A606380F-8A93-4D0C-BF6D-C62C0335E05E@icm.uu.se]
När du har kontakt med oss på Uppsala universitet med e-post så innebär det att vi behandlar dina personuppgifter. För att läsa mer om hur vi gör det kan du läsa här: http://www.uu.se/om-uu/dataskydd-personuppgifter/
E-mailing Uppsala University means that we will process your personal data. For more information on how this is performed, please read here: http://www.uu.se/en/about-uu/data-protection-policy
follow-up: 6 comment:7 by , 4 years ago
Hi Guillame, Thanks! This is great info. I put into the ChimeraX R01 proposal 6 months ago that we will add visualization support for cryoDRGN results and this is a perfect example. I proposed down the road we show the heatmap directly in ChimeraX and let you click any point and ChimeraX will compute the map. (How long are those map computations? I think it executes a neural net so on GPU, how long on a powerful GPU eg. RTX 3090?) But just making an already computed set of maps easy to inspect is obviously a first step. Tom
comment:8 by , 4 years ago
Hi Tom, Yes some keyboard navigation/selection options would be fantastic!! The next/prev/skip interface is a lot better than the model panel, but anything that can minimize mousing while retaining precision and speed would be fantastic. I don't quite know how to convey the use cases better than a screen recording, but I am happy to do whatever I can to help! Have a great weekend Cheers Oli PS It is absolutely absurd to me that Chimera/ChimeraX is having any difficulty whatsoever in getting funding. It is critical structural biology infrastructure, and there are so many improvements to be made with the advent of practical VR/MR, high resolution in situ structural biology, etc. I don't know how funding agencies cannot see this but if there is anything I can do to help (in terms of letters of support from folks here etc) please let me know.
follow-up: 7 comment:9 by , 4 years ago
This type of integration between ChimeraX and cryoDRGN would be so incredibly helpful! As far as I remember, generating these maps doesn’t take much time: a couple minutes on an RTX 2080 Ti for these 50 maps, maybe? I don’t remember exactly, but I also don’t remember having to wait so long as to make me want to switch to another task. So, generating a single map on-demand when the user clicks on the graph probably has a latency well within what’s acceptable for interactive use. If this is helpful to you, I can rerun the generation of these 50 maps next week and time it (and/or time the generation of a single map). I agree with Oli that tools like ChimeraX are absolutely critical. I make sure to cite the ChimeraX paper whenever I publish, but if anything else can help you, let us know. Cheers, Guillaume On 11 Feb 2022, at 20:07, Tom Goddard <goddard@sonic.net<mailto:goddard@sonic.net>> wrote: Hi Guillame, Thanks! This is great info. I put into the ChimeraX R01 proposal 6 months ago that we will add visualization support for cryoDRGN results and this is a perfect example. I proposed down the road we show the heatmap directly in ChimeraX and let you click any point and ChimeraX will compute the map. (How long are those map computations? I think it executes a neural net so on GPU, how long on a powerful GPU eg. RTX 3090?) But just making an already computed set of maps easy to inspect is obviously a first step. Tom När du har kontakt med oss på Uppsala universitet med e-post så innebär det att vi behandlar dina personuppgifter. För att läsa mer om hur vi gör det kan du läsa här: http://www.uu.se/om-uu/dataskydd-personuppgifter/ E-mailing Uppsala University means that we will process your personal data. For more information on how this is performed, please read here: http://www.uu.se/en/about-uu/data-protection-policy
follow-up: 8 comment:10 by , 4 years ago
Showing the cryoDRGN plots and computing the maps on the fly when you click a point is something we proposed in an R01 we are about to submit. If it is funded, that funding won't come through until about a year from now, since that is how long the R01 review and decision cycle takes. But I may work on it sooner. The main trouble is finding time to do it. Do you mean computing 50 maps took a few minutes, or computing each map took a few minutes? And what size were the maps? It will be great if they can be computed in seconds. But even if it is slower we can have ChimeraX compute them in the background and show them when they are ready. Having better controls to visually inspect maps is less work and should be done before the cryoDRGN tool.
follow-up: 9 comment:11 by , 4 years ago
I just ran this again and timed the process: it took 32 seconds to generate these 50 maps, each 128x128x128 voxels, on an RTX 2080 Ti. So well below one second per map. 128x128x128 is the recommended box size for initial assessments of a set of particles with cryoDRGN. Eventually, one would use 256x256x256, but after narrowing down to a much smaller set of particles, so this would require less maps to be sampled from the distribution. Guillaume On 14 Feb 2022, at 19:09, Tom Goddard <goddard@sonic.net<mailto:goddard@sonic.net>> wrote: Showing the cryoDRGN plots and computing the maps on the fly when you click a point is something we proposed in an R01 we are about to submit. If it is funded, that funding won't come through until about a year from now, since that is how long the R01 review and decision cycle takes. But I may work on it sooner. The main trouble is finding time to do it. Do you mean computing 50 maps took a few minutes, or computing each map took a few minutes? And what size were the maps? It will be great if they can be computed in seconds. But even if it is slower we can have ChimeraX compute them in the background and show them when they are ready. Having better controls to visually inspect maps is less work and should be done before the cryoDRGN tool. N?r du har kontakt med oss p? Uppsala universitet med e-post s? inneb?r det att vi behandlar dina personuppgifter. F?r att l?sa mer om hur vi g?r det kan du l?sa h?r: http://www.uu.se/om-uu/dataskydd-personuppgifter/ E-mailing Uppsala University means that we will process your personal data. For more information on how this is performed, please read here: http://www.uu.se/en/about-uu/data-protection-policy
follow-up: 10 comment:12 by , 4 years ago
Hi Guillaume, Wow! That fast compute speed makes me much more interested in trying this calculation soon because it should allow great interactivity exploring the conformational landscape by clicking or dragging a path on the cryoDRGN conformation space plot. Busy submitting two grants right now but let's try this soon. Can you give me some pointers on getting a cryoDRGN test case going? Tom
follow-up: 11 comment:13 by , 4 years ago
Hi Tom, The best starting point is probably the cryoDRGN tutorial: https://www.notion.so/cryoDRGN-tutorial-b932c021cb2c415282f182048bac16ff I am not sure you can download pre-calculated results though. Another option is to re-run one of the analyses from Ellen Zhong’s paper, since she uploaded everything to make it reproducible: https://github.com/zhonge/cryodrgn_empiar The EMPIAR-10076 analysis is a good example of discrete heterogeneity, the EMPIAR-10180 analysis is a good example of continuous heterogeneity. I also have results files that will be part of EMPIAR-10739 when it will be released. I deposited and submitted this entry in September and haven’t heard from EMPIAR since then, despite a couple emails sent to check on the status… not sure what is going on nor when it will be released, but I can definitely send you the files before release. It is the case described in this paper: https://doi.org/10.7554/eLife.71420 Cheers, Guillaume On 15 Feb 2022, at 19:09, Tom Goddard <goddard@sonic.net> wrote: Hi Guillaume, Wow! That fast compute speed makes me much more interested in trying this calculation soon because it should allow great interactivity exploring the conformational landscape by clicking or dragging a path on the cryoDRGN conformation space plot. Busy submitting two grants right now but let's try this soon. Can you give me some pointers on getting a cryoDRGN test case going? Tom När du har kontakt med oss på Uppsala universitet med e-post så innebär det att vi behandlar dina personuppgifter. För att läsa mer om hur vi gör det kan du läsa här: http://www.uu.se/om-uu/dataskydd-personuppgifter/ E-mailing Uppsala University means that we will process your personal data. For more information on how this is performed, please read here: http://www.uu.se/en/about-uu/data-protection-policy
follow-up: 12 comment:14 by , 4 years ago
Thanks for the tutorial and test data info. Realistically this isn't going to get attention until our two main ChimeraX grants are resubmitted by mid-March, and after that we will have a rather enormous backlog of other deferred work. So it will be some time before I can work on cryoDRGN visualization.
Begin forwarded message:
From: Oliver Clarke
Subject: Re: [chimerax-users] Next/previous buttons?
Date: February 7, 2022 at 6:20:48 AM PST
To: Guillaume Gaullier
Cc: Tom Goddard , vincent Chaptal , ChimeraX Users Help <chimerax-users@…>
Hi all,
Here is a quick screen recording:
https://www.dropbox.com/s/jlbhbjqy9mdrb74/screen_recording_next_prev.mov?dl=0
This is a mock up, so there's not much interesting to see, but in this case I am comparing different maps from the output of a 3D classification run with an atomic model. The interface in chimera allows me to easily and quickly compare all the classes to both each other and to the atomic model, identify the good ones and then compare those, while deselecting the others, quickly flick between just two classes, and then to compare two classes with a fixed third class.
Currently I find it hard to do anything like this in ChimeraX, but it may be just that I haven't spent enough time with it.
In "real" examples I would be looking for subtle conformational differences (or perhaps the presence/absence of a ligand/ion/water at higher resolution) between the classes, which flicking quickly between maps can help discern, but there is not much to see on that front here - does this recording make it easier to understand the use case?
Cheers
Oli