Opened 4 years ago
Closed 19 months ago
#5401 closed defect (fixed)
Blast Protein: Ligand Formulas (chemical formulas) hard to read
| Reported by: | Elaine Meng | Owned by: | Zach Pearson |
|---|---|---|---|
| Priority: | blocked | Milestone: | |
| Component: | Sequence | Version: | |
| Keywords: | Cc: | Eric Pettersen | |
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description (last modified by )
The Ligand Formulas (chemical formulas of ligand residues) are hard to read. A single formula contains spaces, so it's hard to parse where one ligand ends and another starts. Also, the numbers should be subscripts if possible.
E.g. the formula of one ligand type (ligand symbol GOL) is currently shown as "C3 H8 O3" ... instead would be better without spaces and with numbers as subscripts like a standard chemical formula. https://en.wikipedia.org/wiki/Chemical_formula
Attachments (2)
Change History (11)
comment:1 by , 4 years ago
comment:2 by , 4 years ago
That work is on hotfix/blast_ligand_formulas when you get a chance to look at it.
comment:3 by , 4 years ago
It looks like getting HTML / rich text to display within a Qt table is a mild nightmare, so I am going to open a new ticket for that, blocking this one. So for now you should take the spaces out of the formula. Whenever I close that other ticket, you could add the subscripting markup here. Very unlikely to make the 1.3 release.
comment:4 by , 4 years ago
| Blocked By: | → 5407 |
|---|
comment:5 by , 4 years ago
| Priority: | moderate → blocked |
|---|
comment:6 by , 3 years ago
I put a QStyledItemDelegate in the blastprotein code for this column, but it doesn't elide -- QTextDocument's CSS engine doesn't handle the text-overflow property. At least this implementation doesn't force all the text over the other columns though. Should we scrap it until we figure out a way to put an ellipse there or should we be OK with clipping the text?
follow-up: 7 comment:7 by , 3 years ago
Hi Zach, I don't know whether the change you mention is already in the daily build, but the problem appears to have been solved already in the daily build UCSF ChimeraX version: 1.6.dev202304050652 (2023-04-05) I.e. in this screenshot from that build, there are no spaces in the chemical formulae of the ligands so I am satisfied with how they look. Also the ellipses are fine and consistent with other columns, so perhaps it would be better to back out whatever you just did.
comment:8 by , 20 months ago
| Blocked By: | 5407 |
|---|---|
| Description: | modified (diff) |
by , 19 months ago
| Attachment: | Screenshot 2024-08-14 at 14.29.56.png added |
|---|
Chemical formulas formatted via HTML
comment:9 by , 19 months ago
| Resolution: | → fixed |
|---|---|
| Status: | assigned → closed |
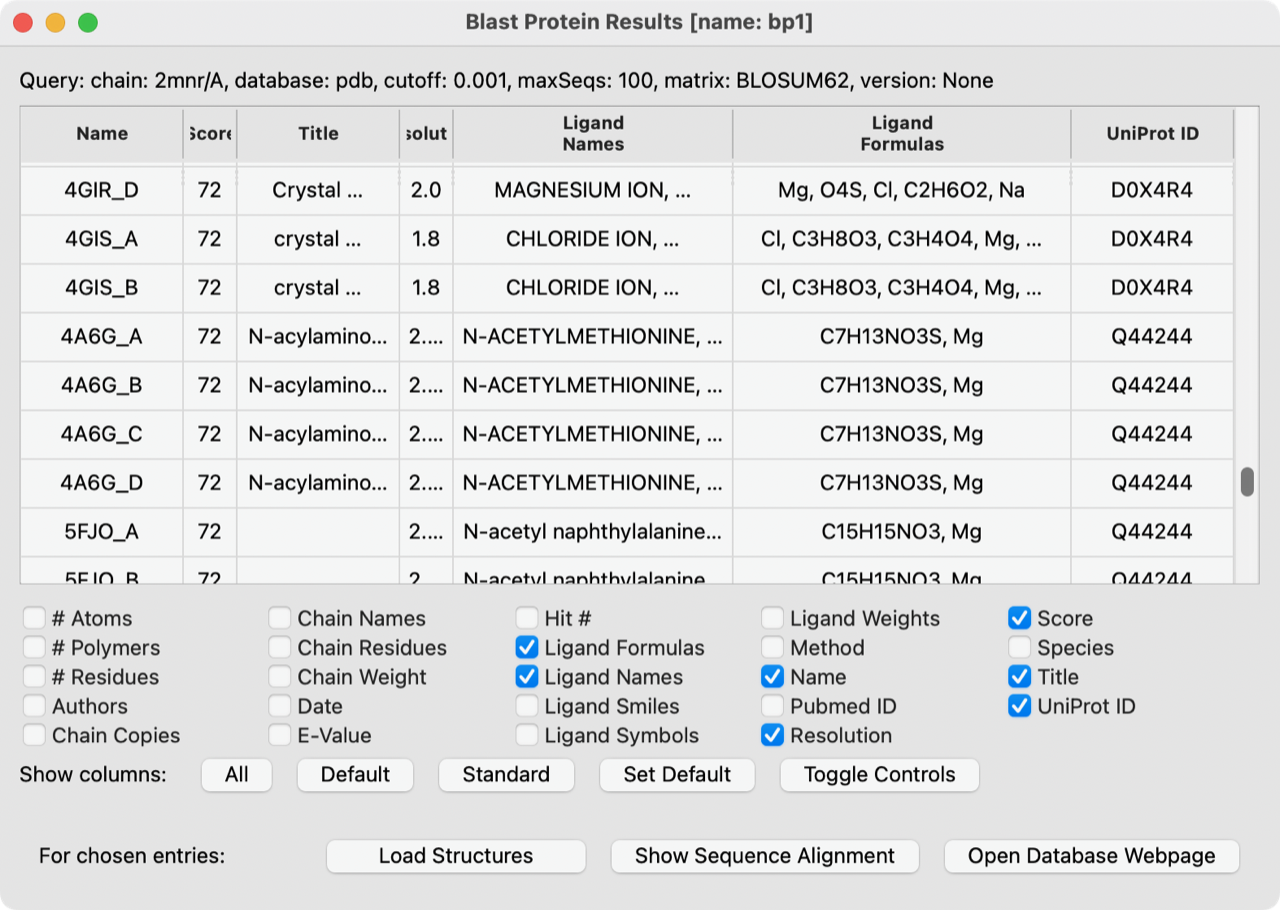
I'm still not sure how to acess the cell widgets in
ItemTable, but I understand that Qt widgets can take HTML so I have modified the data postprocessing logic to change strings likeC8 H15 N O6to
C<sub>8</sub>H<sub>15</sub>NO<sub>6</sub>Can you help out with getting it displayed correctly Eric?