#4483 closed defect (fixed)
side by side camera: zoom misaligns
| Reported by: | Owned by: | Tom Goddard | |
|---|---|---|---|
| Priority: | normal | Milestone: | |
| Component: | Graphics | Version: | |
| Keywords: | Cc: | Tristan Croll | |
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
The following bug report has been submitted:
Platform: Linux-4.15.0-140-generic-x86_64-with-debian-buster-sid
ChimeraX Version: 1.1 (2020-09-09 22:22:27 UTC)
Description
(Describe the actions that caused this problem to occur here)
I tried to use the "camera sbs" side- by-side stereo mode, which I'm pretty sure worked in an earlier version. The problem is that when you zoom in, the two views are horribly misaligned. I think the problem might be that the zoom is centered on the space each image might take if the mode were mono.
Log:
UCSF ChimeraX version: 1.1 (2020-09-09)
© 2016-2020 Regents of the University of California. All rights reserved.
How to cite UCSF ChimeraX
Successfully installed 'ChimeraX-ISOLDE==1.1.0'
Looking in indexes: https://pypi.org/simple,
https://cxtoolshed.rbvi.ucsf.edu/pypi/
Requirement already satisfied: ChimeraX-AddH==2.1.3 in
/home/rjr27/.local/share/ChimeraX/1.1/site-packages (2.1.3)
Requirement already satisfied: ChimeraX-Clipper==0.15.0 in
/home/rjr27/.local/share/ChimeraX/1.1/site-packages (0.15.0)
Requirement already satisfied: ChimeraX-ISOLDE==1.1.0 in
/home/rjr27/.local/share/ChimeraX/1.1/site-packages (1.1.0)
Requirement already satisfied: ChimeraX-mmCIF~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (2.2)
Requirement already satisfied: ChimeraX-Hbonds~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-AtomSearch~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-Atomic~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.6.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Geometry~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Atomic~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.6.1)
Requirement already satisfied: ChimeraX-Arrays~=1.0.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Clipper==0.15.0) (1.0)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Atomic~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.6.1)
Requirement already satisfied: ChimeraX-Clipper==0.15.0 in
/home/rjr27/.local/share/ChimeraX/1.1/site-packages (0.15.0)
Requirement already satisfied: ChimeraX-Arrays~=1.0.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Clipper==0.15.0) (1.0)
Requirement already satisfied: ChimeraX-Atomic~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.6.1)
Requirement already satisfied: ChimeraX-PDB~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (2.1)
Requirement already satisfied: ChimeraX-SaveCommand~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (1.2)
Requirement already satisfied: ChimeraX-OpenCommand~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (1.2.1)
Requirement already satisfied: ChimeraX-DataFormats~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-Arrays~=1.0.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Clipper==0.15.0) (1.0)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-AtomSearch~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-UI~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Hbonds~=2.0->ChimeraX-
AddH==2.1.3) (1.2.3)
Requirement already satisfied: ChimeraX-Atomic~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.6.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-ChemGroup~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Hbonds~=2.0->ChimeraX-
AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-Geometry~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Atomic~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.6.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-mmCIF~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (2.2)
Requirement already satisfied: ChimeraX-PDB~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (2.1)
Requirement already satisfied: ChimeraX-Graphics~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Atomic~=1.0->ChimeraX-
AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-ConnectStructure~=2.0 in
/usr/lib/ucsf-chimerax/lib/python3.7/site-packages (from ChimeraX-
Atomic~=1.0->ChimeraX-AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-Arrays~=1.0.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Clipper==0.15.0) (1.0)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Nucleotides~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Atomic~=1.0->ChimeraX-
AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-Geometry~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Arrays~=1.0.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Clipper==0.15.0) (1.0)
Requirement already satisfied: ChimeraX-SaveCommand~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (1.2)
Requirement already satisfied: ChimeraX-OpenCommand~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (1.2.1)
Requirement already satisfied: ChimeraX-DataFormats~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-Arrays~=1.0.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Clipper==0.15.0) (1.0)
Requirement already satisfied: ChimeraX-Atomic~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.6.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-IO~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-
SaveCommand~=1.0->ChimeraX-mmCIF~=2.0->ChimeraX-AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-DataFormats~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-IO~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-
SaveCommand~=1.0->ChimeraX-mmCIF~=2.0->ChimeraX-AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-UI~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Hbonds~=2.0->ChimeraX-
AddH==2.1.3) (1.2.3)
Requirement already satisfied: ChimeraX-DataFormats~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-OpenCommand~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (1.2.1)
Requirement already satisfied: ChimeraX-IO~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-
SaveCommand~=1.0->ChimeraX-mmCIF~=2.0->ChimeraX-AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-SaveCommand~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (1.2)
Requirement already satisfied: ChimeraX-Atomic~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.6.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Arrays~=1.0.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Clipper==0.15.0) (1.0)
Requirement already satisfied: ChimeraX-Geometry~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Arrays~=1.0.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Clipper==0.15.0) (1.0)
Requirement already satisfied: ChimeraX-Atomic~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.6.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-AtomSearch~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-Surface~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-
Nucleotides~=2.0->ChimeraX-Atomic~=1.0->ChimeraX-AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-UI~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Hbonds~=2.0->ChimeraX-
AddH==2.1.3) (1.2.3)
Requirement already satisfied: ChimeraX-Atomic~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.6.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Geometry~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-OpenCommand~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (1.2.1)
Requirement already satisfied: ChimeraX-Graphics~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Atomic~=1.0->ChimeraX-
AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-DataFormats~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-Arrays~=1.0.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Clipper==0.15.0) (1.0)
Requirement already satisfied: ChimeraX-StdCommands~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Surface~=1.0->ChimeraX-
Nucleotides~=2.0->ChimeraX-Atomic~=1.0->ChimeraX-AddH==2.1.3) (1.0.4)
Requirement already satisfied: ChimeraX-MapData~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Surface~=1.0->ChimeraX-
Nucleotides~=2.0->ChimeraX-Atomic~=1.0->ChimeraX-AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-Map~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Surface~=1.0->ChimeraX-
Nucleotides~=2.0->ChimeraX-Atomic~=1.0->ChimeraX-AddH==2.1.3) (1.0.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Geometry~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-mmCIF~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (2.2)
Requirement already satisfied: ChimeraX-Graphics~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Atomic~=1.0->ChimeraX-
AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-Atomic~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.6.1)
Requirement already satisfied: ChimeraX-Dssp~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-
StdCommands~=1.0->ChimeraX-Surface~=1.0->ChimeraX-Nucleotides~=2.0->ChimeraX-
Atomic~=1.0->ChimeraX-AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-MapFit~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-
StdCommands~=1.0->ChimeraX-Surface~=1.0->ChimeraX-Nucleotides~=2.0->ChimeraX-
Atomic~=1.0->ChimeraX-AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Geometry~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-DataFormats~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-Geometry~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-MouseModes~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Map~=1.0->ChimeraX-
Surface~=1.0->ChimeraX-Nucleotides~=2.0->ChimeraX-Atomic~=1.0->ChimeraX-
AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-SaveCommand~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (1.2)
Requirement already satisfied: ChimeraX-OpenCommand~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-mmCIF~=2.0->ChimeraX-
AddH==2.1.3) (1.2.1)
Requirement already satisfied: ChimeraX-Graphics~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Atomic~=1.0->ChimeraX-
AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-MapSeries~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Map~=1.0->ChimeraX-
Surface~=1.0->ChimeraX-Nucleotides~=2.0->ChimeraX-Atomic~=1.0->ChimeraX-
AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-UI~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Hbonds~=2.0->ChimeraX-
AddH==2.1.3) (1.2.3)
Requirement already satisfied: ChimeraX-Arrays~=1.0.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Clipper==0.15.0) (1.0)
Requirement already satisfied: ChimeraX-MapData~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Surface~=1.0->ChimeraX-
Nucleotides~=2.0->ChimeraX-Atomic~=1.0->ChimeraX-AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-MapFilter~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Map~=1.0->ChimeraX-
Surface~=1.0->ChimeraX-Nucleotides~=2.0->ChimeraX-Atomic~=1.0->ChimeraX-
AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-Geometry~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Atomic~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.6.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Arrays~=1.0.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Clipper==0.15.0) (1.0)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-MapData~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Surface~=1.0->ChimeraX-
Nucleotides~=2.0->ChimeraX-Atomic~=1.0->ChimeraX-AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-Map~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Surface~=1.0->ChimeraX-
Nucleotides~=2.0->ChimeraX-Atomic~=1.0->ChimeraX-AddH==2.1.3) (1.0.1)
Requirement already satisfied: ChimeraX-Geometry~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Graphics~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Atomic~=1.0->ChimeraX-
AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-Geometry~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-UI~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Hbonds~=2.0->ChimeraX-
AddH==2.1.3) (1.2.3)
Requirement already satisfied: ChimeraX-MapData~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Surface~=1.0->ChimeraX-
Nucleotides~=2.0->ChimeraX-Atomic~=1.0->ChimeraX-AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-MapFit~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-
StdCommands~=1.0->ChimeraX-Surface~=1.0->ChimeraX-Nucleotides~=2.0->ChimeraX-
Atomic~=1.0->ChimeraX-AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-Map~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Surface~=1.0->ChimeraX-
Nucleotides~=2.0->ChimeraX-Atomic~=1.0->ChimeraX-AddH==2.1.3) (1.0.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Geometry~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-MapData~=2.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Surface~=1.0->ChimeraX-
Nucleotides~=2.0->ChimeraX-Atomic~=1.0->ChimeraX-AddH==2.1.3) (2.0)
Requirement already satisfied: ChimeraX-Map~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Surface~=1.0->ChimeraX-
Nucleotides~=2.0->ChimeraX-Atomic~=1.0->ChimeraX-AddH==2.1.3) (1.0.1)
Requirement already satisfied: ChimeraX-Geometry~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-MouseModes~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Map~=1.0->ChimeraX-
Surface~=1.0->ChimeraX-Nucleotides~=2.0->ChimeraX-Atomic~=1.0->ChimeraX-
AddH==2.1.3) (1.0)
Requirement already satisfied: ChimeraX-Core~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-AddH==2.1.3) (1.1)
Requirement already satisfied: ChimeraX-Graphics~=1.0 in /usr/lib/ucsf-
chimerax/lib/python3.7/site-packages (from ChimeraX-Atomic~=1.0->ChimeraX-
AddH==2.1.3) (1.0)
Lock 140491113182352 acquired on
/home/rjr27/.cache/ChimeraX/1.1/toolshed/bundle_info.cache.lock
Lock 140491113182352 released on
/home/rjr27/.cache/ChimeraX/1.1/toolshed/bundle_info.cache.lock
WARNING: You are using pip version 20.2.2; however, version 21.0.1 is
available.
You should consider upgrading via the '/usr/bin/chimerax -m pip install
--upgrade pip' command.
> open /home/rjr27/phaser/MR/JasonGuo/20191125_A1B10BR2_ML195_refine_182.pdb
Summary of feedback from opening
/home/rjr27/phaser/MR/JasonGuo/20191125_A1B10BR2_ML195_refine_182.pdb
---
warnings | Ignored bad PDB record found on line 1
REMARK Date 2021-03-19 Time 09:29:15 GMT +0000 (1616146155.93 s)
Ignored bad PDB record found on line 2
REMARK PHENIX refinement
Ignored bad PDB record found on line 4
REMARK ****************** INPUT FILES AND LABELS
******************************
Ignored bad PDB record found on line 5
REMARK Reflections:
Ignored bad PDB record found on line 6
REMARK file name :
/Users/Jason/DLS/20191125/ALK1B10BR2L/ML195/Phenix/bFuLefOLZXOYfheV-SWS-
merged-aniso.mtz
46 messages similar to the above omitted
Chain information for 20191125_A1B10BR2_ML195_refine_182.pdb #1
---
Chain | Description
A B C D | No description available
E | No description available
F G H | No description available
I | No description available
J | No description available
K | No description available
L | No description available
> ui tool show ISOLDE
> set selectionWidth 4
Chain information for 20191125_A1B10BR2_ML195_refine_182.pdb
---
Chain | Description
1.2/A 1.2/B 1.2/C 1.2/D | No description available
1.2/E | No description available
1.2/F 1.2/G 1.2/H | No description available
1.2/I | No description available
1.2/J | No description available
1.2/K | No description available
1.2/L | No description available
Cached rota8000-val data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-leu data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-ile data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-pro data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-phe data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-tyr data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-trp data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-ser data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-thr data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-cys data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-met data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-lys data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-his data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-arg data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-asp data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-asn data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-gln data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rota8000-glu data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rama8000-cispro data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rama8000-transpro data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rama8000-gly-sym data not found. Regenerating from text file. This is
normal if running ISOLDE for the first time
Cached rama8000-prepro-noGP data not found. Regenerating from text file. This
is normal if running ISOLDE for the first time
Cached rama8000-ileval-nopreP data not found. Regenerating from text file.
This is normal if running ISOLDE for the first time
Cached rama8000-general-noGPIVpreP data not found. Regenerating from text
file. This is normal if running ISOLDE for the first time
Forcefield cache not found or out of date. Regenerating from ffXML files. This
is normal if running ISOLDE for the first time, or after upgrading OpenMM.
Done loading forcefield
> set bgColor white
> view #1/F:87
Reflection data provided as intensities. Performing French & Wilson scaling to
convert to amplitudes...
> camera sbs
OpenGL version: 3.3.0 NVIDIA 440.33.01
OpenGL renderer: Quadro RTX 5000/PCIe/SSE2
OpenGL vendor: NVIDIA Corporation
Manufacturer: Dell Inc.
Model: Precision 5820 Tower X-Series
OS: Ubuntu 18.04 bionic
Architecture: 64bit ELF
CPU: 16 Intel(R) Core(TM) i7-9800X CPU @ 3.80GHz
Cache Size: 16896 KB
Memory:
total used free shared buff/cache available
Mem: 125G 3.1G 97G 300M 24G 121G
Swap: 15G 0B 15G
Graphics:
65:00.0 VGA compatible controller [0300]: NVIDIA Corporation Device [10de:1eb0] (rev a1)
Subsystem: Dell Device [1028:129f]
Kernel driver in use: nvidia
PyQt version: 5.12.3
Compiled Qt version: 5.12.4
Runtime Qt version: 5.12.9
Installed Packages:
alabaster: 0.7.12
appdirs: 1.4.4
Babel: 2.8.0
backcall: 0.2.0
blockdiag: 2.0.1
certifi: 2020.6.20
chardet: 3.0.4
ChimeraX-AddH: 2.1.3
ChimeraX-AlignmentAlgorithms: 2.0
ChimeraX-AlignmentHdrs: 3.2
ChimeraX-AlignmentMatrices: 2.0
ChimeraX-Alignments: 2.1
ChimeraX-Arrays: 1.0
ChimeraX-Atomic: 1.6.1
ChimeraX-AtomSearch: 2.0
ChimeraX-AxesPlanes: 2.0
ChimeraX-BasicActions: 1.1
ChimeraX-BILD: 1.0
ChimeraX-BlastProtein: 1.0.1
ChimeraX-BondRot: 2.0
ChimeraX-BugReporter: 1.0
ChimeraX-BuildStructure: 2.0
ChimeraX-Bumps: 1.0
ChimeraX-BundleBuilder: 1.0
ChimeraX-ButtonPanel: 1.0
ChimeraX-CageBuilder: 1.0
ChimeraX-CellPack: 1.0
ChimeraX-Centroids: 1.1
ChimeraX-ChemGroup: 2.0
ChimeraX-Clashes: 2.0
ChimeraX-Clipper: 0.15.0
ChimeraX-ColorActions: 1.0
ChimeraX-ColorGlobe: 1.0
ChimeraX-CommandLine: 1.1.3
ChimeraX-ConnectStructure: 2.0
ChimeraX-Contacts: 1.0
ChimeraX-Core: 1.1
ChimeraX-CoreFormats: 1.0
ChimeraX-coulombic: 1.0.1
ChimeraX-Crosslinks: 1.0
ChimeraX-Crystal: 1.0
ChimeraX-DataFormats: 1.0
ChimeraX-Dicom: 1.0
ChimeraX-DistMonitor: 1.1
ChimeraX-DistUI: 1.0
ChimeraX-Dssp: 2.0
ChimeraX-EMDB-SFF: 1.0
ChimeraX-ExperimentalCommands: 1.0
ChimeraX-FileHistory: 1.0
ChimeraX-FunctionKey: 1.0
ChimeraX-Geometry: 1.1
ChimeraX-gltf: 1.0
ChimeraX-Graphics: 1.0
ChimeraX-Hbonds: 2.0
ChimeraX-Help: 1.0
ChimeraX-HKCage: 1.0
ChimeraX-IHM: 1.0
ChimeraX-ImageFormats: 1.0
ChimeraX-IMOD: 1.0
ChimeraX-IO: 1.0
ChimeraX-ISOLDE: 1.1.0
ChimeraX-Label: 1.0
ChimeraX-LinuxSupport: 1.0
ChimeraX-ListInfo: 1.0
ChimeraX-Log: 1.1.1
ChimeraX-LookingGlass: 1.1
ChimeraX-Map: 1.0.1
ChimeraX-MapData: 2.0
ChimeraX-MapEraser: 1.0
ChimeraX-MapFilter: 2.0
ChimeraX-MapFit: 2.0
ChimeraX-MapSeries: 2.0
ChimeraX-Markers: 1.0
ChimeraX-Mask: 1.0
ChimeraX-MatchMaker: 1.1
ChimeraX-MDcrds: 2.0
ChimeraX-MedicalToolbar: 1.0.1
ChimeraX-Meeting: 1.0
ChimeraX-MLP: 1.0
ChimeraX-mmCIF: 2.2
ChimeraX-MMTF: 2.0
ChimeraX-Modeller: 1.0
ChimeraX-ModelPanel: 1.0
ChimeraX-ModelSeries: 1.0
ChimeraX-Mol2: 2.0
ChimeraX-Morph: 1.0
ChimeraX-MouseModes: 1.0
ChimeraX-Movie: 1.0
ChimeraX-Neuron: 1.0
ChimeraX-Nucleotides: 2.0
ChimeraX-OpenCommand: 1.2.1
ChimeraX-PDB: 2.1
ChimeraX-PDBBio: 1.0
ChimeraX-PickBlobs: 1.0
ChimeraX-Positions: 1.0
ChimeraX-PresetMgr: 1.0
ChimeraX-PubChem: 2.0
ChimeraX-Read-Pbonds: 1.0
ChimeraX-Registration: 1.1
ChimeraX-RemoteControl: 1.0
ChimeraX-ResidueFit: 1.0
ChimeraX-RestServer: 1.0
ChimeraX-RNALayout: 1.0
ChimeraX-RotamerLibMgr: 2.0
ChimeraX-RotamerLibsDunbrack: 2.0
ChimeraX-RotamerLibsDynameomics: 2.0
ChimeraX-RotamerLibsRichardson: 2.0
ChimeraX-SaveCommand: 1.2
ChimeraX-SchemeMgr: 1.0
ChimeraX-SDF: 2.0
ChimeraX-Segger: 1.0
ChimeraX-Segment: 1.0
ChimeraX-SeqView: 2.2
ChimeraX-Shape: 1.0.1
ChimeraX-Shell: 1.0
ChimeraX-Shortcuts: 1.0
ChimeraX-ShowAttr: 1.0
ChimeraX-ShowSequences: 1.0
ChimeraX-SideView: 1.0
ChimeraX-Smiles: 2.0
ChimeraX-SmoothLines: 1.0
ChimeraX-SpaceNavigator: 1.0
ChimeraX-StdCommands: 1.0.4
ChimeraX-STL: 1.0
ChimeraX-Storm: 1.0
ChimeraX-Struts: 1.0
ChimeraX-Surface: 1.0
ChimeraX-SwapAA: 2.0
ChimeraX-SwapRes: 2.0
ChimeraX-TapeMeasure: 1.0
ChimeraX-Test: 1.0
ChimeraX-Toolbar: 1.0
ChimeraX-ToolshedUtils: 1.0
ChimeraX-Tug: 1.0
ChimeraX-UI: 1.2.3
ChimeraX-uniprot: 2.0
ChimeraX-ViewDockX: 1.0
ChimeraX-Vive: 1.1
ChimeraX-VolumeMenu: 1.0
ChimeraX-VTK: 1.0
ChimeraX-WavefrontOBJ: 1.0
ChimeraX-WebCam: 1.0
ChimeraX-WebServices: 1.0
ChimeraX-Zone: 1.0
colorama: 0.4.3
comtypes: 1.1.7
cxservices: 1.0
cycler: 0.10.0
Cython: 0.29.20
decorator: 4.4.2
distlib: 0.3.1
distro: 1.5.0
docutils: 0.16
filelock: 3.0.12
funcparserlib: 0.3.6
grako: 3.16.5
h5py: 2.10.0
html2text: 2020.1.16
idna: 2.10
ihm: 0.16
imagecodecs: 2020.5.30
imagecodecs-lite: 2020.1.31
imagesize: 1.2.0
ipykernel: 5.3.0
ipython: 7.15.0
ipython-genutils: 0.2.0
jedi: 0.17.2
Jinja2: 2.11.2
jupyter-client: 6.1.3
jupyter-core: 4.6.3
kiwisolver: 1.2.0
line-profiler: 2.1.2
lxml: 4.5.1
MarkupSafe: 1.1.1
matplotlib: 3.2.1
msgpack: 1.0.0
netifaces: 0.10.9
networkx: 2.4
numexpr: 2.7.1
numpy: 1.18.5
numpydoc: 1.0.0
openvr: 1.12.501
packaging: 20.4
parso: 0.7.1
pexpect: 4.8.0
pickleshare: 0.7.5
Pillow: 7.1.2
pip: 20.2.2
pkginfo: 1.5.0.1
prompt-toolkit: 3.0.7
psutil: 5.7.0
ptyprocess: 0.6.0
pycollada: 0.7.1
pydicom: 2.0.0
Pygments: 2.6.1
PyOpenGL: 3.1.5
PyOpenGL-accelerate: 3.1.5
pyparsing: 2.4.7
PyQt5-commercial: 5.12.3
PyQt5-sip: 4.19.19
PyQtWebEngine-commercial: 5.12.1
python-dateutil: 2.8.1
pytz: 2020.1
pyzmq: 19.0.2
qtconsole: 4.7.4
QtPy: 1.9.0
RandomWords: 0.3.0
requests: 2.24.0
scipy: 1.4.1
setuptools: 49.4.0
sfftk-rw: 0.6.6.dev0
six: 1.15.0
snowballstemmer: 2.0.0
sortedcontainers: 2.2.2
Sphinx: 3.1.1
sphinxcontrib-applehelp: 1.0.2
sphinxcontrib-blockdiag: 2.0.0
sphinxcontrib-devhelp: 1.0.2
sphinxcontrib-htmlhelp: 1.0.3
sphinxcontrib-jsmath: 1.0.1
sphinxcontrib-qthelp: 1.0.3
sphinxcontrib-serializinghtml: 1.1.4
suds-jurko: 0.6
tables: 3.6.1
tifffile: 2020.6.3
tinyarray: 1.2.2
tornado: 6.0.4
traitlets: 5.0.4
urllib3: 1.25.10
wcwidth: 0.2.5
webcolors: 1.11.1
wheel: 0.34.2
Attachments (1)
Change History (11)
comment:1 by , 5 years ago
| Component: | Unassigned → Graphics |
|---|---|
| Owner: | set to |
| Platform: | → all |
| Project: | → ChimeraX |
| Status: | new → assigned |
| Summary: | ChimeraX bug report submission → side by side camera: zoom misaligns |
comment:2 by , 5 years ago
| Cc: | added |
|---|
I tried camera sbs with zooming in ChimeraX 1.0, 1.1 and daily build and all worked the same and seemed to work correctly to me. I judged it by looking cross-eyed at the structure and zooming made it bigger and smaller without moving it closer or further from view which seems correct.
My suspicion is that ISOLDE changes the zoom mode and that is what make the side-by-side zoom not work right. I will try that later today after several meetings.
follow-up: 3 comment:3 by , 5 years ago
That certainly sounds likely! I'm afraid none of ISOLDE's mouse modes are tested in stereo.
________________________________
From: ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu>
Sent: 08 April 2021 18:08
Cc: goddard@cgl.ucsf.edu <goddard@cgl.ucsf.edu>; Randy John Read <rjr27@cam.ac.uk>; Tristan Croll <tic20@cam.ac.uk>
Subject: Re: [ChimeraX] #4483: side by side camera: zoom misaligns
#4483: side by side camera: zoom misaligns
-------------------------------+-------------------------
Reporter: rjr27@… | Owner: Tom Goddard
Type: defect | Status: assigned
Priority: normal | Milestone:
Component: Graphics | Version:
Resolution: | Keywords:
Blocked By: | Blocking:
Notify when closed: | Platform: all
Project: ChimeraX |
-------------------------------+-------------------------
Changes (by Tom Goddard):
* cc: Tristan Croll (added)
Comment:
I tried camera sbs with zooming in ChimeraX 1.0, 1.1 and daily build and
all worked the same and seemed to work correctly to me. I judged it by
looking cross-eyed at the structure and zooming made it bigger and smaller
without moving it closer or further from view which seems correct.
My suspicion is that ISOLDE changes the zoom mode and that is what make
the side-by-side zoom not work right. I will try that later today after
several meetings.
--
Ticket URL: <https://plato.cgl.ucsf.edu/trac/ChimeraX/ticket/4483#comment:2>
ChimeraX <http://www.rbvi.ucsf.edu/chimerax/>
ChimeraX Issue Tracker
follow-up: 4 comment:4 by , 5 years ago
No, I’m afraid this happens even without having loaded ISOLDE, so we can’t blame Tristan!
I can reproduce it with the following (ChimeraX 1.1, both on a Mac and a Linux box running Ubuntu 18.04):
Launch ChimeraX, then give the following commands:
open 1sgt
view #1/A:246
clip off
camera sbs
(The “clip off” command is because hardly anything is visible after the view command, which is centered on the single Ca atom in this structure.)
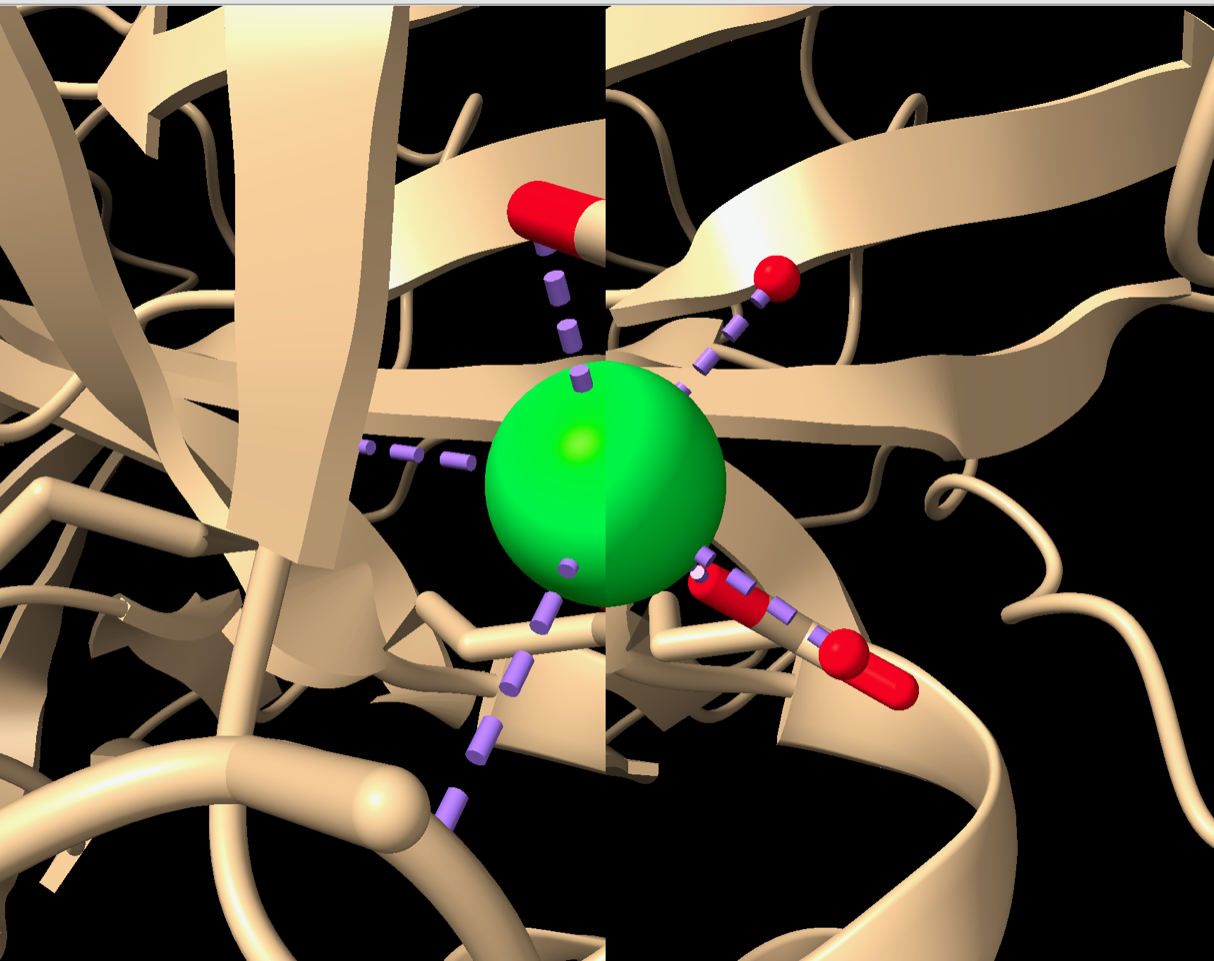
After this I rotated the view so the Ca atom was at the front and gradually zoomed in. When zoomed out or at intermediate zooms it seems fine, but as you zoom in closer the Ca atom (at the center of the viewport) moves closer and closer to the center line dividing the two views, until you get what is in the screenshot: the left image only shows what is to the left of the Ca atom and the right image only shows what is to the right, leaving nothing in common to give stereo perception. I’m guessing you didn’t zoom in close enough to notice it degrading.
Best wishes,
Randy
[cid:4FD99EC2-AF73-49D9-8CE2-790067AEE1F7@cable.virginm.net]
On 8 Apr 2021, at 19:25, ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu<mailto:ChimeraX-bugs-admin@cgl.ucsf.edu>> wrote:
#4483: side by side camera: zoom misaligns
-------------------------------+-------------------------
Reporter: rjr27@… | Owner: Tom Goddard
Type: defect | Status: assigned
Priority: normal | Milestone:
Component: Graphics | Version:
Resolution: | Keywords:
Blocked By: | Blocking:
Notify when closed: | Platform: all
Project: ChimeraX |
-------------------------------+-------------------------
Comment (by Tristan Croll):
{{{
That certainly sounds likely! I'm afraid none of ISOLDE's mouse modes are
tested in stereo.
________________________________
From: ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu<mailto:ChimeraX-bugs-admin@cgl.ucsf.edu>>
Sent: 08 April 2021 18:08
Cc: goddard@cgl.ucsf.edu<mailto:goddard@cgl.ucsf.edu> <goddard@cgl.ucsf.edu<mailto:goddard@cgl.ucsf.edu>>; Randy John Read
<rjr27@cam.ac.uk<mailto:rjr27@cam.ac.uk>>; Tristan Croll <tic20@cam.ac.uk<mailto:tic20@cam.ac.uk>>
Subject: Re: [ChimeraX] #4483: side by side camera: zoom misaligns
#4483: side by side camera: zoom misaligns
-------------------------------+-------------------------
Reporter: rjr27@… | Owner: Tom Goddard
Type: defect | Status: assigned
Priority: normal | Milestone:
Component: Graphics | Version:
Resolution: | Keywords:
Blocked By: | Blocking:
Notify when closed: | Platform: all
Project: ChimeraX |
-------------------------------+-------------------------
Changes (by Tom Goddard):
* cc: Tristan Croll (added)
Comment:
I tried camera sbs with zooming in ChimeraX 1.0, 1.1 and daily build and
all worked the same and seemed to work correctly to me. I judged it by
looking cross-eyed at the structure and zooming made it bigger and
smaller
without moving it closer or further from view which seems correct.
My suspicion is that ISOLDE changes the zoom mode and that is what make
the side-by-side zoom not work right. I will try that later today after
several meetings.
--
Ticket URL:
<https://plato.cgl.ucsf.edu/trac/ChimeraX/ticket/4483#comment:2>
ChimeraX <http://www.rbvi.ucsf.edu/chimerax/>
ChimeraX Issue Tracker
}}}
--
Ticket URL: <https://plato.cgl.ucsf.edu/trac/ChimeraX/ticket/4483#comment:3>
ChimeraX <http://www.rbvi.ucsf.edu/chimerax/>
ChimeraX Issue Tracker
-----
Randy J. Read
Department of Haematology, University of Cambridge
Cambridge Institute for Medical Research Tel: +44 1223 336500
The Keith Peters Building Fax: +44 1223 336827
Hills Road E-mail: rjr27@cam.ac.uk<mailto:rjr27@cam.ac.uk>
Cambridge CB2 0XY, U.K. www-structmed.cimr.cam.ac.uk<http://www-structmed.cimr.cam.ac.uk>
comment:5 by , 5 years ago
Thanks! Sorry for blaming Tristan! This specific test case will help me figure it out. You say some older ChimeraX worked? Because I don't think the side-by-side stereo has been changed, but I can try very old ChimeraX and see. I am afraid the wrong behavior is because side-by-side stereo is *not* cross-eye or wall-eye stereo. It is meant for 3D displays that take as input two parallel camera views -- ie both eye views have their center direction exactly parallel (not converging). There is no cross-eye or wall-eye in ChimeraX although it could be added.
follow-up: 5 comment:6 by , 5 years ago
Hi Tom, It’s quite possible that I tried the camera sbs mode before but never zoomed in enough to notice that something was wrong. It’s also possible that I’m one of only a very small number of users who would really like to have wall-eyed split-screen stereo, but it’s something I’ve used a lot, starting way back with the MMS-X graphics machine in the late 1970s! Best wishes, Randy ----- Randy J. Read Department of Haematology, University of Cambridge Cambridge Institute for Medical Research Tel: +44 1223 336500 The Keith Peters Building Fax: +44 1223 336827 Hills Road E-mail: rjr27@cam.ac.uk Cambridge CB2 0XY, U.K. www-structmed.cimr.cam.ac.uk
follow-up: 6 comment:7 by , 5 years ago
| Resolution: | → fixed |
|---|---|
| Status: | assigned → closed |
Hi Randy,
Ok, zooming in side-by-side camera mode will be fixed in tomorrow's ChimeraX daily build (about 5 AM California time, afternoon in Cambridge). The bug was that there are two ways to zoom, one is to move the camera (view point) closer to the models, and the other is to scale the models to a larger size. For non-stereo viewing these appear the same. But for stereo they don't have the same effect because in the first mode the eye separation stays the same distance relative to model size, while in the second mode the eye separation is reduced relative to the model size. Stereo needs that second mode. ChimeraX was correctly using that for sequential stereo mode (LCD shutter glasses) but not for the side-by-side stereo mode. I made it use the correct type of zooming for side-by-side and now it works fine to zoom in close.
Also I said before that side-by-side is not the same as wall-eye, but I think that is wrong. Side-by-side mode is exactly wall-eye as long as you make your window width on the screen twice your physical eye spacing. For cross-eye the side-by-side mode is not good -- first the two views have to be swapped, but also the size of the two views on your screen can be large and knowing that size relative to your eye spacing is important to getting correct depth scale. So cross-eye mode really needs another parameter.
Thanks for pointing out this bug. Let me know if it needs more work -- I don't use wall-eye much but I remember the people who love it at UCSF would tell me that some software did wall-eye much better than others, and Chimera was supposedly not good at it.
I do the ChimeraX VR development and have been trying to get Tristan Croll interested enough to get a VR headset for ISOLDE. I think VR can be an excellent addition for solving X-ray and cryoEM structures. A few years ago Bob Stroud came to try ChimeraX VR and we looked at a structure he published in Science the month before. Within 30 seconds he said "What is going on with arginine 149? It can't be pointing in that direction in the binding site." He then had his post-doc who was with us look and she agreed - the structure was wrong. How many months did they look at this on the flat screen and not see the problem? Matt Jacobson who works on drug binding used VR every week before the pandemic -- super expert at drug binding -- and I remember him standing in a binding site in VR at his group meeting saying "I didn't realize there was all this room over here in the binding site." This has happened so many times that experts at flat-screen structure visualization have commented how obvious things were in VR that they never realized on the flat screen despite decades of flat-screen 3D experience. Anyways, that is my pitch for trying VR.
Tom
comment:8 by , 5 years ago
Hi Tom, Thanks for fixing it so quickly and thanks for the clear explanation! Tristan can of course speak for himself, but he was expressing interest in exploring VR and, as I recall, had just arranged to visit someone with a good VR setup to test it out when Covid-19 came around and we were all locked down! So far my only experience with current VR technology was 5 minutes with CootVR, where it became obvious that the difficult part is to reimagine the way you interact with the model. The CootVR interface was terribly clunky and unintuitive. When we’re back in the lab it would be good to check out how VR in ChimeraX works. We’ve also talked a bit about the potential for AR, where you could possibly have more intuitive interactions like pulling on an atom with your fingers. But I’m sure that getting that to feel right would be quite a challenge! Randy ----- Randy J. Read Department of Haematology, University of Cambridge Cambridge Institute for Medical Research Tel: +44 1223 336500 The Keith Peters Building Fax: +44 1223 336827 Hills Road E-mail: rjr27@cam.ac.uk Cambridge CB2 0XY, U.K. www-structmed.cimr.cam.ac.uk
follow-up: 8 comment:9 by , 5 years ago
Randy's pretty much covered it, but my own two cents: given the pace at which VR technology has advanced, I've felt it would be silly to go ahead and buy a headset until I'm sure I'll have the time and space to properly use it - the last thing I want is to leave it gathering dust as it rapidly goes obsolete. I talked to Randy about buying one at the start of 2020 - but with the lockdown it quickly became clear that I wasn't going to have much coding time for quite a while. The last year has borne that out - even if I'd bought the headset, ISOLDE-VR would most likely still have only seen a few days of real development.
A thought, now that it appears a return to relative normality is in sight: perhaps this could be part of my proposed contribution to the ChimeraX R01?
________________________________
From: ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu>
Sent: 09 April 2021 08:56
To: Randy John Read <rjr27@cam.ac.uk>; goddard@cgl.ucsf.edu <goddard@cgl.ucsf.edu>
Cc: Tristan Croll <tic20@cam.ac.uk>
Subject: Re: [ChimeraX] #4483: side by side camera: zoom misaligns
#4483: side by side camera: zoom misaligns
-------------------------------+-------------------------
Reporter: rjr27@… | Owner: Tom Goddard
Type: defect | Status: closed
Priority: normal | Milestone:
Component: Graphics | Version:
Resolution: fixed | Keywords:
Blocked By: | Blocking:
Notify when closed: | Platform: all
Project: ChimeraX |
-------------------------------+-------------------------
Comment (by rjr27@…):
{{{
Hi Tom,
Thanks for fixing it so quickly and thanks for the clear explanation!
Tristan can of course speak for himself, but he was expressing interest in
exploring VR and, as I recall, had just arranged to visit someone with a
good VR setup to test it out when Covid-19 came around and we were all
locked down!
So far my only experience with current VR technology was 5 minutes with
CootVR, where it became obvious that the difficult part is to reimagine
the way you interact with the model. The CootVR interface was terribly
clunky and unintuitive. When we’re back in the lab it would be good to
check out how VR in ChimeraX works.
We’ve also talked a bit about the potential for AR, where you could
possibly have more intuitive interactions like pulling on an atom with
your fingers. But I’m sure that getting that to feel right would be quite
a challenge!
Randy
-----
Randy J. Read
Department of Haematology, University of Cambridge
Cambridge Institute for Medical Research Tel: +44 1223 336500
The Keith Peters Building Fax: +44 1223
336827
Hills Road E-mail:
rjr27@cam.ac.uk
Cambridge CB2 0XY, U.K. www-
structmed.cimr.cam.ac.uk
}}}
--
Ticket URL: <https://plato.cgl.ucsf.edu/trac/ChimeraX/ticket/4483#comment:8>
ChimeraX <http://www.rbvi.ucsf.edu/chimerax/>
ChimeraX Issue Tracker
follow-up: 9 comment:10 by , 5 years ago
Hi Randy, Tristan, Yeah, I reviewed the CootVR manuscript. User interface that just uses the VR hand controllers and no keyboard or mouse is a big stumbling block. VR definitely isn't useful for structure building yet, but I think with a good bit of work it could be and it would allow higher quality structures, more problems caught and fixed. I totally understand Tristan that you have too much to do right now. I think it is my job to make the VR work well in ISOLDE, not yours. The VR headsets are pretty cheap and the hardware is not evolving as fast as you think. An Oculus Quest 2 with link cable to a PC is about $400. At the high end a Valve Index is $1000. While I agree it is wasteful to get equipment when you have no time right now to use it, it still could make some sense to get it. Because when you do get a little time here and there and want to work on something new and fun, if you don't have the equipment it won't happen, while if it is sitting on your desk you may make some progress. These VR headsets should be good for about 2-3 more years. Currently we don't plan on putting any VR into the new ChimeraX R01 proposal. We do not have it in the current R01 either. VR is funded by a separate contract from NIAID and we would have problems with overlapping funding if we added it to the R01. Tom
Reported by Randy Read