Opened 17 months ago
Last modified 17 months ago
#15998 feedback defect
Cannot update file history
| Reported by: | Owned by: | Eric Pettersen | |
|---|---|---|---|
| Priority: | normal | Milestone: | |
| Component: | UI | Version: | |
| Keywords: | Cc: | chimera-programmers | |
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
The following bug report has been submitted:
Platform: Windows-10-10.0.22631
ChimeraX Version: 1.8 (2024-06-10 23:15:52 UTC)
Description
Hello, I'm having issues saving a .cxs session. After saving, the log says I have permissions denied. I've tried un-installing and re-installing several versiosn of ChimeraX over the past few months and nothing has resolved this saving issue. This is the pop-up window notifying failure to save:
"Error processing trigger "file history changed":
PermissionError: [Errno 13] Permission denied: 'C:\\Users\\chris\\AppData\\Local\\Temp\\chbp8qmgij_1.html'
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-packages\chimerax\ui\widgets\htmlview.py", line 244, in setHtml
tf = open(self._tf_name, "wb")
^^^^^^^^^^^^^^^^^^^^^^^^^"
I attached a note file of the log text.
Thanks for the help.
Log:
UCSF ChimeraX version: 1.8 (2024-06-10)
© 2016-2024 Regents of the University of California. All rights reserved.
> surface cap true
Log from Wed Sep 4 19:17:24 2024UCSF ChimeraX version: 1.6.1 (2023-05-09)
© 2016-2023 Regents of the University of California. All rights reserved.
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Supplemental Figure #_modes of neut/9-4-2024_Gc with all 4
> Sites models.cxs"
Opened cryosparc_P409_J122_004_volume_map_sharp.mrc as #1, grid size
220,220,220, pixel 1.88, shown at level 0.2, step 1, values float32
Log from Thu Aug 15 18:02:26 2024UCSF ChimeraX version: 1.6.1 (2023-05-09)
© 2016-2023 Regents of the University of California. All rights reserved.
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site 6/8-15-2024_Site 6 models and J122 map for
> figure.cxs"
Opened cryosparc_P409_J122_004_volume_map_sharp.mrc as #1, grid size
220,220,220, pixel 1.88, shown at level 0.2, step 1, values float32
Log from Thu Aug 15 11:55:54 2024 Startup Messages
---
warning | Replacing fetcher for 'pdb_nmr' and format NMRSTAR from NMRSTAR bundle with that from NMRSTAR bundle
UCSF ChimeraX version: 1.6.1 (2023-05-09)
© 2016-2023 Regents of the University of California. All rights reserved.
How to cite UCSF ChimeraX
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Structure Files/Gc-4Ffabs/Finalizing Site 6 cryo
> map/cryosparc_P409_J122_004_volume_map_sharp.mrc"
Opened cryosparc_P409_J122_004_volume_map_sharp.mrc as #1, grid size
220,220,220, pixel 1.88, shown at level 0.0904, step 1, values float32
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Structure Files/Gc-4Ffabs/Finalizing Site 6 cryo
> map/cryosparc_P409_J125_003_volume_map_sharp.mrc"
Opened cryosparc_P409_J125_003_volume_map_sharp.mrc as #2, grid size
220,220,220, pixel 1.88, shown at level 0.0551, step 1, values float32
> open
> C:/Users/ckh755/Downloads/cryosparc_P409_J120_class_01_00062_volume_sharp.mrc
Opened cryosparc_P409_J120_class_01_00062_volume_sharp.mrc as #3, grid size
128,128,128, pixel 3.23, shown at level 0.0946, step 1, values float32
> open
> C:/Users/ckh755/Downloads/cryosparc_P409_J120_class_01_00062_volume_sharp.mrc
Opened cryosparc_P409_J120_class_01_00062_volume_sharp.mrc as #4, grid size
128,128,128, pixel 3.23, shown at level 0.0946, step 1, values float32
> open C:/Users/ckh755/Downloads/cryosparc_P409_J119_class_01_final_volume.mrc
Opened cryosparc_P409_J119_class_01_final_volume.mrc as #5, grid size
192,192,192, pixel 2.15, shown at level 0.0663, step 1, values float32
> open C:/Users/ckh755/Downloads/cryosparc_P409_J111_003_volume_map_sharp.mrc
Opened cryosparc_P409_J111_003_volume_map_sharp.mrc as #6, grid size
220,220,220, pixel 1.88, shown at level 0.05, step 1, values float32
> close #4
> open C:/Users/ckh755/Downloads/cryosparc_P409_J105_004_volume_map_sharp.mrc
Opened cryosparc_P409_J105_004_volume_map_sharp.mrc as #4, grid size
220,220,220, pixel 1.88, shown at level 0.0941, step 1, values float32
> preset "overall look" "publication 1 (silhouettes)"
Using preset: Overall Look / Publication 1 (Silhouettes)
Preset expands to these ChimeraX commands:
set bg white
graphics silhouettes t
lighting depthCue f
> vop step 1
> vop level 0.2
> surface dust #1 size 18.8
> surface dust #2 size 18.8
> surface dust #3 size 32.3
> surface dust #5 size 21.5
> surface dust #6 size 18.8
> surface dust #4 size 18.8
> hide #!6 models
> hide #!5 models
> hide #!4 models
> hide #!3 models
> hide #!2 models
> hide #!1 models
> show #!2 models
> show #!1 models
> show #!3 models
> hide #!2 models
> hide #!1 models
> hide #!3 models
> show #!4 models
> hide #!4 models
> show #!5 models
> show #!6 models
> hide #!5 models
> close #2-6
> show #!1 models
> volume #1 level 0.21
> volume #1 level 0.2
> surface dust #1 size 18.8
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site
> 6/Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH.pdb"
Chain information for
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH.pdb #2
---
Chain | Description
A | No description available
B | No description available
G | No description available
H | No description available
L | No description available
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site
> 6/Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH.pdb"
Chain information for
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH.pdb #3
---
Chain | Description
A | No description available
B | No description available
G | No description available
H | No description available
L | No description available
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site
> 6/Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH.pdb"
Chain information for
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH.pdb #4
---
Chain | Description
A | No description available
B | No description available
G | No description available
H | No description available
L | No description available
> rename #2 Gc-42437-36124_37847refined_350-noH.pdb
> hide #!3 models
> hide #!1 models
> hide #!4 models
> close #2-4
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site
> 6/Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for
> 37847 Fv model.pdb"
Chain information for
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for 37847
Fv model.pdb #2
---
Chain | Description
A | No description available
B | No description available
G | No description available
H | No description available
L | No description available
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site
> 6/Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for
> 42437 Fv model.pdb"
Chain information for
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for 42437
Fv model.pdb #3
---
Chain | Description
A | No description available
B | No description available
G | No description available
H | No description available
L | No description available
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site
> 6/Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
> domain I-II model.pdb"
Chain information for
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
domain I-II model.pdb #4
---
Chain | Description
A | No description available
B | No description available
G | No description available
H | No description available
L | No description available
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site 6/Gc-42437-36124_refine_035_for 36124 Fab
> model.pdb"
Chain information for Gc-42437-36124_refine_035_for 36124 Fab model.pdb #5
---
Chain | Description
A | No description available
B | No description available
G | No description available
H | No description available
L | No description available
> mmaker #5 to #2
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for 37847
Fv model.pdb, chain G (#2) with Gc-42437-36124_refine_035_for 36124 Fab
model.pdb, chain G (#5), sequence alignment score = 1511.8
RMSD between 235 pruned atom pairs is 0.897 angstroms; (across all 321 pairs:
2.280)
> hide #2-5 target a
> cartoon #2-5
> hide #!5 models
> show #!5 models
> hide #!5 models
> show #!5 models
> hide #!5 models
> hide #!4 models
> hide #!3 models
Drag select of 150 residues
> select up
2125 atoms, 2171 bonds, 271 residues, 1 model selected
> select up
4240 atoms, 4343 bonds, 547 residues, 1 model selected
> cartoon hide sel
> hide #!2 models
> show #!3 models
Drag select of 227 residues, 1 pseudobonds
> select up
2634 atoms, 2700 bonds, 1 pseudobond, 331 residues, 2 models selected
> select up
4264 atoms, 4371 bonds, 1 pseudobond, 546 residues, 2 models selected
> cartoon hide (#!3 & sel)
> hide #!3 models
> show #!4 models
> select clear
Drag select of 61 residues
> select up
834 atoms, 850 bonds, 110 residues, 1 model selected
> select up
1714 atoms, 1756 bonds, 226 residues, 1 model selected
> cartoon hide sel
Drag select of 143 residues
> select up
1440 atoms, 1472 bonds, 186 residues, 1 model selected
> select up
1738 atoms, 1784 bonds, 225 residues, 1 model selected
> cartoon hide sel
> show #!5 models
> hide #!4 models
Drag select of 92 residues
> select up
1185 atoms, 1206 bonds, 156 residues, 1 model selected
> select up
1714 atoms, 1754 bonds, 226 residues, 1 model selected
> cartoon hide sel
> show #!4 models
> hide #!4 models
> show #!4 models
> hide #!4 models
> show #!4 models
> hide #!4 models
> show #!4 models
> hide #!4 models
> show #!4 models
> show #!1 models
> select add #1
1714 atoms, 1754 bonds, 226 residues, 3 models selected
> select add #5
8882 atoms, 9094 bonds, 1 pseudobond, 1154 residues, 4 models selected
> select subtract #5
2 models selected
> ui mousemode right "translate selected models"
> ui mousemode right "rotate selected models"
> view matrix models
> #1,-0.98262,-0.1739,-0.064914,446.81,-0.063593,0.64393,-0.76244,239.77,0.17439,-0.74506,-0.6438,449.76
> view matrix models
> #1,0.99758,-0.031402,-0.061997,19.397,-0.025974,-0.99591,0.086502,359.5,-0.06446,-0.084682,-0.99432,448.57
> ui mousemode right "translate selected models"
> view matrix models
> #1,0.99758,-0.031402,-0.061997,19.753,-0.025974,-0.99591,0.086502,394.2,-0.06446,-0.084682,-0.99432,442.7
> view matrix models
> #1,0.99758,-0.031402,-0.061997,21.992,-0.025974,-0.99591,0.086502,395.32,-0.06446,-0.084682,-0.99432,437.53
> select subtract #1
Nothing selected
> transparency #1,4-5 50
> ui tool show "Fit in Map"
> fitmap #4 inMap #1
Fit molecule
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
domain I-II model.pdb (#4) to map cryosparc_P409_J122_004_volume_map_sharp.mrc
(#1) using 5978 atoms
average map value = 0.3436, steps = 88
shifted from previous position = 8.78
rotated from previous position = 7.03 degrees
atoms outside contour = 1389, contour level = 0.2
Position of
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
domain I-II model.pdb (#4) relative to
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) coordinates:
Matrix rotation and translation
0.99414132 0.07879143 -0.07399286 0.45674897
0.07681640 -0.99661836 -0.02917344 400.23012853
-0.07604126 0.02331866 -0.99683196 426.55477183
Axis 0.99853400 0.03896580 -0.03757008
Axis point 0.00000000 197.29859251 215.92025858
Rotation angle (degrees) 178.49383096
Shift along axis 0.02566740
> fitmap #5 inMap #1
Fit molecule Gc-42437-36124_refine_035_for 36124 Fab model.pdb (#5) to map
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) using 8882 atoms
average map value = 0.2656, steps = 80
shifted from previous position = 4.74
rotated from previous position = 9.22 degrees
atoms outside contour = 3338, contour level = 0.2
Position of Gc-42437-36124_refine_035_for 36124 Fab model.pdb (#5) relative to
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) coordinates:
Matrix rotation and translation
-0.08397317 -0.06990450 0.99401301 188.88436047
-0.45519181 0.89006610 0.02414025 143.00806885
-0.88642480 -0.45043945 -0.10656163 111.56904090
Axis -0.24001391 0.95101250 -0.19485518
Axis point 168.97538130 0.00000000 -22.39729916
Rotation angle (degrees) 98.64050778
Shift along axis 68.92778219
> hide #!1 models
> show #!1 models
> hide #!5 models
> hide #!4 models
> show #!5 models
> show #!4 models
> hide #!5 models
> show #!5 models
> hide #!5 models
> show #!5 models
> hide #!4 models
> hide #!1 models
Drag select of 61 residues
> select up
1021 atoms, 1034 bonds, 129 residues, 1 model selected
> select up
3756 atoms, 3845 bonds, 481 residues, 1 model selected
> cartoon hide sel
> show #!4 models
> select add #5
8882 atoms, 9094 bonds, 1 pseudobond, 1154 residues, 2 models selected
> show #!3 models
> show #!2 models
> show #!1 models
> select subtract #5
Nothing selected
> hide #!5 models
> hide #!4 models
> hide #!3 models
> hide #!2 models
> hide #!1 models
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Structure Files/Gc-4Ffabs/Map for Gc245 med res map with Site 6 and
> domain III/fold_gc_diii_36145_fv_model_4.cif"
Chain information for fold_gc_diii_36145_fv_model_4.cif #6
---
Chain | Description
A | .
B | .
C | .
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Structure Files/Gc+Site2+Site5/Pymol/Gc-42437-36124_refine_035.pdb"
Chain information for Gc-42437-36124_refine_035.pdb #7
---
Chain | Description
A | No description available
B | No description available
G | No description available
H | No description available
L | No description available
> mmaker #6 to #7
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker Gc-42437-36124_refine_035.pdb, chain G (#7) with
fold_gc_diii_36145_fv_model_4.cif, chain A (#6), sequence alignment score =
520.8
RMSD between 105 pruned atom pairs is 0.570 angstroms; (across all 105 pairs:
0.570)
> hide #6-7 target a
> select add #7
8882 atoms, 9094 bonds, 1 pseudobond, 1154 residues, 2 models selected
> show #7 cartoons
> select subtract #7
Nothing selected
> show #!4 models
> hide #!4 models
> close #6#7
> show #!5 models
> show #!4 models
> show #!3 models
> show #!2 models
> show #!1 models
> vop step 1
> vop level 0.2
> hide dust
Expected a collection of one of 'atoms', 'bonds', 'cartoons', 'models',
'pbonds', 'pseudobonds', 'ribbons', or 'surfaces' or a keyword
> close #2-5
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site 6/Models for
> Figure/Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for
> Gc domain I-II model.pdb"
Chain information for
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
domain I-II model.pdb #2
---
Chain | Description
A | No description available
B | No description available
G | No description available
H | No description available
L | No description available
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site 6/Models for
> Figure/fold_gc_diii_36145_fv_model_4.cif"
Chain information for fold_gc_diii_36145_fv_model_4.cif #3
---
Chain | Description
A | .
B | .
C | .
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site 6/Models for
> Figure/Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for
> 42437 Fv model.pdb"
Chain information for
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for 42437
Fv model.pdb #4
---
Chain | Description
A | No description available
B | No description available
G | No description available
H | No description available
L | No description available
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site 6/Models for
> Figure/Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for
> 37847 Fv model.pdb"
Chain information for
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for 37847
Fv model.pdb #5
---
Chain | Description
A | No description available
B | No description available
G | No description available
H | No description available
L | No description available
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site 6/Models for
> Figure/Gc-42437-36124_refine_035_for 36124 Fab model.pdb"
Chain information for Gc-42437-36124_refine_035_for 36124 Fab model.pdb #6
---
Chain | Description
A | No description available
B | No description available
G | No description available
H | No description available
L | No description available
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site 6/Models for
> Figure/fold_gc_diii_36145_fv_model_4.cif"
Chain information for fold_gc_diii_36145_fv_model_4.cif #7
---
Chain | Description
A | .
B | .
C | .
> mmaker #3-7 too #2
> matchmaker #3-7 too #2
Expected a keyword
> mmaker #3-7 to #2
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
domain I-II model.pdb, chain L (#2) with fold_gc_diii_36145_fv_model_4.cif,
chain C (#3), sequence alignment score = 509
RMSD between 106 pruned atom pairs is 0.629 angstroms; (across all 108 pairs:
0.736)
Matchmaker
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
domain I-II model.pdb, chain G (#2) with
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for 42437
Fv model.pdb, chain G (#4), sequence alignment score = 1686.8
RMSD between 321 pruned atom pairs is 0.000 angstroms; (across all 321 pairs:
0.000)
Matchmaker
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
domain I-II model.pdb, chain G (#2) with
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for 37847
Fv model.pdb, chain G (#5), sequence alignment score = 1686.8
RMSD between 321 pruned atom pairs is 0.000 angstroms; (across all 321 pairs:
0.000)
Matchmaker
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
domain I-II model.pdb, chain G (#2) with Gc-42437-36124_refine_035_for 36124
Fab model.pdb, chain G (#6), sequence alignment score = 1511.8
RMSD between 235 pruned atom pairs is 0.897 angstroms; (across all 321 pairs:
2.280)
Matchmaker
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
domain I-II model.pdb, chain L (#2) with fold_gc_diii_36145_fv_model_4.cif,
chain C (#7), sequence alignment score = 509
RMSD between 106 pruned atom pairs is 0.629 angstroms; (across all 108 pairs:
0.736)
> hide #3 models
> hide #!4 models
> hide #!5 models
> hide #!6 models
> hide #7 models
> hide #!2 models
> show #3 models
> show #!6 models
> hide #3 models
> show #!5 models
> show #7 models
> show #!4 models
> show #3 models
> show #!2 models
> mmaker #3,7 to #6
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker Gc-42437-36124_refine_035_for 36124 Fab model.pdb, chain G (#6)
with fold_gc_diii_36145_fv_model_4.cif, chain A (#3), sequence alignment score
= 520.8
RMSD between 105 pruned atom pairs is 0.570 angstroms; (across all 105 pairs:
0.570)
Matchmaker Gc-42437-36124_refine_035_for 36124 Fab model.pdb, chain G (#6)
with fold_gc_diii_36145_fv_model_4.cif, chain A (#7), sequence alignment score
= 520.8
RMSD between 105 pruned atom pairs is 0.570 angstroms; (across all 105 pairs:
0.570)
> hide atoms
> show cartoons
> hide #7 models
> hide #!6 models
> hide #!5 models
> hide #!4 models
> hide #3 models
> hide #!1 models
> hide #2/H,L,A,B
> hide cartoons #2/H,L,A,B
Expected ',' or a keyword
> hide cartoons #2/H,L,A,B
Expected ',' or a keyword
> hide #2/H,L,A,B cartoons
[Repeated 1 time(s)]
> show #3 models
> hide #!2 models
> show #!4 models
> hide #3 models
> show #!5 models
> show #!2 models
> hide #4/G,A,B cartoons
> hide #5/G,H,L cartoons
> hide #!5 models
> hide #!4 models
> hide #!2 models
> show #!2 models
> show #!6 models
> hide #!2 models
> hide #6/G,H,L cartoons
> show #!5 models
> show #!4 models
> show #3 models
> hide #3 models
> show #!2 models
> hide #!4 models
> hide #!2 models
> hide #!6 models
> show #!4 models
> hide #!5 models
> show #!6 models
> hide #!4 models
> hide #!6 models
> show #7 models
> show #3 models
> show #!2 models
> hide #7 models
> hide #3 models
> hide #!2 models
> show #7 models
> show #!6 models
> show #!5 models
> show #!4 models
> show #3 models
> show #!2 models
> color #3/B,C #DD833E
> color #7/B,C #DD833E
> color #6/A,B #78C14D
> color #5/A,B #CC66A7
> color #4/H,L #9142AE
> dssp
> color #2/G:1041-1140,1232-1268,1419-1430 #FF7F7F
[Repeated 1 time(s)]
> color
> #2/G:1141-1186,1205-1231,1269-1282,1296-1320,1347-1403,1410-1418,1187-1204
> #FFFF7F
> color #2/G:1283-1295,1321-1346,1404-1409 #c18c7f
> color #2/G:1442-1556, #7F7FFF
Expected a color or one of 'byatom', 'bychain', 'byelement', 'byhetero',
'byidentity', 'bymodel', 'bynucleotide', 'bypolymer', 'fromatoms',
'fromcartoons', 'fromribbons', or 'random' or a keyword
> color #2/G:1431-1441, #7FFFFF
Expected a color or one of 'byatom', 'bychain', 'byelement', 'byhetero',
'byidentity', 'bymodel', 'bynucleotide', 'bypolymer', 'fromatoms',
'fromcartoons', 'fromribbons', or 'random' or a keyword
> color #2/G:1431-1441, #7FFFFF
Expected a color or one of 'byatom', 'bychain', 'byelement', 'byhetero',
'byidentity', 'bymodel', 'bynucleotide', 'bypolymer', 'fromatoms',
'fromcartoons', 'fromribbons', or 'random' or a keyword
> color #2/G:1442-1556 #7F7FFF
> color #2/G:1431-1441 #7FFFFF
> color #3/A:1442-1556 #7F7FFF
> color #7/A:1442-1556 #7F7FFF
> hide #!2 models
> hide #!4 models
> hide #!5 models
> hide #!6 models
> show #!2 models
> hide #!2 models
> color #3/A:1442-1556 #7F7FFF
> color #7/A:1442-1556 #7F7FFF
Drag select of 74 residues
> select up
922 atoms, 934 bonds, 124 residues, 2 models selected
> select up
1592 atoms, 1616 bonds, 210 residues, 2 models selected
> color sel #7F7FFF
> ui tool show "Show Sequence Viewer"
> sequence chain #3/A
Alignment identifier is 3/A
> color sel green
> color #3/A:1-105 #7F7FFF
> color #7/A:1-105 #7F7FFF
> show #!6 models
> show #!5 models
> show #!4 models
> show #!2 models
> show #!1 models
> select clear
> hide #!1 models
> show
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
> sticks
Expected a collection of one of 'atoms', 'bonds', 'cartoons', 'models',
'pbonds', 'pseudobonds', 'ribbons', or 'surfaces' or a keyword
> color
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
> #33FF33
> undo
> color
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
> #33FF33
> select
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
72 atoms, 67 bonds, 12 residues, 1 model selected
> show sel atoms
> show sel atoms sticks
Expected ',' or a keyword
> show sel atoms as sticks
Expected ',' or a keyword
> style sel stick
Changed 72 atom styles
> color
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
> #33FF33
> save "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site 6/8-15-2024_Site 6 models and J122 map for
> figure.cxs" includeMaps true
——— End of log from Thu Aug 15 11:55:54 2024 ———
opened ChimeraX session
> select
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
> sidechain only
Expected a keyword
> select sidechain only
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
Expected a keyword
> select sidechain
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
Expected a keyword
> color
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
> sideonly
Expected a color or one of 'byatom', 'bychain', 'byelement', 'byhetero',
'byidentity', 'bymodel', 'bynucleotide', 'bypolymer', 'fromatoms',
'fromcartoons', 'fromribbons', or 'random' or a keyword
> color
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
> sideonly #33FF33
Expected a color or one of 'byatom', 'bychain', 'byelement', 'byhetero',
'byidentity', 'bymodel', 'bynucleotide', 'bypolymer', 'fromatoms',
'fromcartoons', 'fromribbons', or 'random' or a keyword
> select clear
> select
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
> sideonly
Expected a keyword
> select
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
> sidechainonly
Expected a keyword
> select
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
> sidechain
Expected a keyword
> select sidechain
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
Expected a keyword
> select side
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
Expected an objects specifier or a keyword
> select sideonly
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
Expected a keyword
> select
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
> sideonly
Expected a keyword
> select
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
72 atoms, 67 bonds, 12 residues, 1 model selected
> show sel atoms as sticks
Expected ',' or a keyword
> show (sel-residues & sidechain) target ab
> select clear
> select
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
72 atoms, 67 bonds, 12 residues, 1 model selected
> select #3/A:Cys
48 atoms, 40 bonds, 8 residues, 1 model selected
> select #3,7/A:Cys
96 atoms, 80 bonds, 16 residues, 2 models selected
> select #3/A:Cys
48 atoms, 40 bonds, 8 residues, 1 model selected
> select #3,7/A:Cys
96 atoms, 80 bonds, 16 residues, 2 models selected
> show (sel-residues & sidechain) target ab
> show sel target ab
> dssp
> select up
572 atoms, 574 bonds, 78 residues, 2 models selected
> select up
1592 atoms, 1616 bonds, 210 residues, 2 models selected
> select up
5098 atoms, 5204 bonds, 662 residues, 2 models selected
> select clear
> select #3,7/A:Cys
96 atoms, 80 bonds, 16 residues, 2 models selected
> bond sel
Created 8 bonds
> select
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
72 atoms, 67 bonds, 12 residues, 1 model selected
> select #3,7/A:Cys@ca
16 atoms, 16 residues, 2 models selected
> select #3,7/A:Cys@c
16 atoms, 16 residues, 2 models selected
> select #3,7/A:Cys@sc
Nothing selected
> select #3,7/A:Cys@S
Nothing selected
> select #3,7/A:Cys target a
Expected a keyword
> color #3,7/A:Cys orange target a
> color #3,7/A:Cys #33FF33 target a
> color
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
> #33FF33 target a
> select clear
> show #2-7 target ab
> cartoon #2-7
> hide #2-7 target a
> color #2 #dadadaff
> color #3 #dadadaff
> color #4 #dadadaff
> color #5 #dadadaff
> color #6 #dadadaff
> color #7 #dadadaff
> dssp
> mmaker #3-7 to #2
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
domain I-II model.pdb, chain L (#2) with fold_gc_diii_36145_fv_model_4.cif,
chain C (#3), sequence alignment score = 509
RMSD between 106 pruned atom pairs is 0.629 angstroms; (across all 108 pairs:
0.736)
Matchmaker
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
domain I-II model.pdb, chain G (#2) with
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for 42437
Fv model.pdb, chain G (#4), sequence alignment score = 1686.8
RMSD between 321 pruned atom pairs is 0.000 angstroms; (across all 321 pairs:
0.000)
Matchmaker
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
domain I-II model.pdb, chain G (#2) with
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for 37847
Fv model.pdb, chain G (#5), sequence alignment score = 1686.8
RMSD between 321 pruned atom pairs is 0.000 angstroms; (across all 321 pairs:
0.000)
Matchmaker
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
domain I-II model.pdb, chain G (#2) with Gc-42437-36124_refine_035_for 36124
Fab model.pdb, chain G (#6), sequence alignment score = 1511.8
RMSD between 235 pruned atom pairs is 0.897 angstroms; (across all 321 pairs:
2.280)
Matchmaker
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
domain I-II model.pdb, chain L (#2) with fold_gc_diii_36145_fv_model_4.cif,
chain C (#7), sequence alignment score = 509
RMSD between 106 pruned atom pairs is 0.629 angstroms; (across all 108 pairs:
0.736)
> mmaker #3,7 to #6
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker Gc-42437-36124_refine_035_for 36124 Fab model.pdb, chain G (#6)
with fold_gc_diii_36145_fv_model_4.cif, chain A (#3), sequence alignment score
= 520.8
RMSD between 105 pruned atom pairs is 0.570 angstroms; (across all 105 pairs:
0.570)
Matchmaker Gc-42437-36124_refine_035_for 36124 Fab model.pdb, chain G (#6)
with fold_gc_diii_36145_fv_model_4.cif, chain A (#7), sequence alignment score
= 520.8
RMSD between 105 pruned atom pairs is 0.570 angstroms; (across all 105 pairs:
0.570)
> hide atoms
> show cartoons
> hide #2/H,L,A,B cartoons
> hide #4/G,A,B cartoons
> hide #5/G,H,L cartoons
> hide #6/G,H,L cartoons
> color #4/H,L #9142AE
> color #5/A,B #CC66A7
> color #6/A,B #78C14D
> color #3/B,C #DD833E
> color #7/B,C #DD833E
> color #2/G:1041-1140,1232-1268,1419-1430 #FF7F7F
> color
> #2/G:1141-1186,1205-1231,1269-1282,1296-1320,1347-1403,1410-1418,1187-1204
> #FFFF7F
> color #2/G:1283-1295,1321-1346,1404-1409 #c18c7f
> color #2/G:1442-1556 #7F7FFF
> color #2/G:1431-1441 #7FFFFF
> color #3/A:1-105 #7F7FFF
> color #7/A:1-105 #7F7FFF
> select
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
72 atoms, 67 bonds, 12 residues, 1 model selected
> show (sel-residues & sidechain) target ab
> color
> #2/G:1165,1198,1169,1205,1173,1207,1175,1180,1193,1360,1208,1368,1236,1246,1305,1310,1452,1464,1455,1458,1519,1539,1528,1531
> #33FF33 target a
> select #3,7/A:Cys
96 atoms, 88 bonds, 16 residues, 2 models selected
> show (sel-residues & sidechain) target ab
> bond sel
Created 0 bonds
> color #3,7/A:Cys #33FF33 target a
> select clear
> show #!1 models
> color #1 #ffffff80 models
> color #1 #ffffff
> transparency #1 0.5
> transparency #1 0.1
> transparency #1 0.9
> hide #!1 models
> show #!1 models
> set trans #1 0.1
Expected a keyword
> set transparent #1 0.1
Expected a keyword
> set transparent #1 0.1
Expected a keyword
> transparency #1 80
> transparency #1 60
> ui tool show "Fit in Map"
> fitmap #2 inMap #1
Fit molecule
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
domain I-II model.pdb (#2) to map cryosparc_P409_J122_004_volume_map_sharp.mrc
(#1) using 5978 atoms
average map value = 0.3436, steps = 88
shifted from previous position = 8.78
rotated from previous position = 7.03 degrees
atoms outside contour = 1389, contour level = 0.2
Position of
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for Gc
domain I-II model.pdb (#2) relative to
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) coordinates:
Matrix rotation and translation
0.99414132 0.07879143 -0.07399286 0.45674897
0.07681640 -0.99661836 -0.02917344 400.23012854
-0.07604126 0.02331866 -0.99683196 426.55477183
Axis 0.99853400 0.03896580 -0.03757008
Axis point 0.00000000 197.29859251 215.92025858
Rotation angle (degrees) 178.49383096
Shift along axis 0.02566740
> fitmap #3 inMap #1
Fit molecule fold_gc_diii_36145_fv_model_4.cif (#3) to map
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) using 2549 atoms
average map value = 0.2661, steps = 236
shifted from previous position = 39.5
rotated from previous position = 85 degrees
atoms outside contour = 977, contour level = 0.2
Position of fold_gc_diii_36145_fv_model_4.cif (#3) relative to
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) coordinates:
Matrix rotation and translation
-0.43327688 -0.58616308 0.68460498 201.78701269
-0.66399689 -0.30601667 -0.68224770 181.73470487
0.60940895 -0.75017774 -0.25662052 177.31430354
Axis -0.53162842 0.58849294 -0.60913650
Axis point 29.96085983 172.73517154 0.00000000
Rotation angle (degrees) 176.33695573
Shift along axis -108.33473362
> fitmap #4 inMap #1
Fit molecule
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for 42437
Fv model.pdb (#4) to map cryosparc_P409_J122_004_volume_map_sharp.mrc (#1)
using 5978 atoms
average map value = 0.3436, steps = 88
shifted from previous position = 8.78
rotated from previous position = 7.03 degrees
atoms outside contour = 1389, contour level = 0.2
Position of
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for 42437
Fv model.pdb (#4) relative to cryosparc_P409_J122_004_volume_map_sharp.mrc
(#1) coordinates:
Matrix rotation and translation
0.99414132 0.07879143 -0.07399286 0.45674897
0.07681640 -0.99661836 -0.02917344 400.23012854
-0.07604126 0.02331866 -0.99683196 426.55477183
Axis 0.99853400 0.03896580 -0.03757008
Axis point 0.00000000 197.29859251 215.92025858
Rotation angle (degrees) 178.49383096
Shift along axis 0.02566740
> fitmap #5 inMap #1
Fit molecule
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for 37847
Fv model.pdb (#5) to map cryosparc_P409_J122_004_volume_map_sharp.mrc (#1)
using 5978 atoms
average map value = 0.3436, steps = 88
shifted from previous position = 8.78
rotated from previous position = 7.03 degrees
atoms outside contour = 1389, contour level = 0.2
Position of
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for 37847
Fv model.pdb (#5) relative to cryosparc_P409_J122_004_volume_map_sharp.mrc
(#1) coordinates:
Matrix rotation and translation
0.99414132 0.07879143 -0.07399286 0.45674897
0.07681640 -0.99661836 -0.02917344 400.23012854
-0.07604126 0.02331866 -0.99683196 426.55477183
Axis 0.99853400 0.03896580 -0.03757008
Axis point 0.00000000 197.29859251 215.92025858
Rotation angle (degrees) 178.49383096
Shift along axis 0.02566740
> fitmap #6 inMap #1
Fit molecule Gc-42437-36124_refine_035_for 36124 Fab model.pdb (#6) to map
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) using 8882 atoms
average map value = 0.2656, steps = 80
shifted from previous position = 4.74
rotated from previous position = 9.22 degrees
atoms outside contour = 3338, contour level = 0.2
Position of Gc-42437-36124_refine_035_for 36124 Fab model.pdb (#6) relative to
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) coordinates:
Matrix rotation and translation
-0.08397309 -0.06990458 0.99401301 188.88436973
-0.45519258 0.89006571 0.02414027 143.00789182
-0.88642441 -0.45044021 -0.10656162 111.56919155
Axis -0.24001431 0.95101233 -0.19485553
Axis point 168.97544778 0.00000000 -22.39719531
Rotation angle (degrees) 98.64051649
Shift along axis 68.92744241
> select add #7
2549 atoms, 2606 bonds, 331 residues, 1 model selected
> select add #3
5098 atoms, 5212 bonds, 662 residues, 2 models selected
> ui mousemode right "rotate selected models"
> view matrix models
> #3,0.8449,-0.50304,0.18194,221.78,0.18775,-0.03963,-0.98142,242.71,0.5009,0.86336,0.060961,268.04,#7,0.8449,-0.50304,0.18194,221.78,0.18775,-0.03963,-0.98142,242.71,0.5009,0.86336,0.060961,268.04
> ui mousemode right "translate selected models"
> view matrix models
> #3,0.8449,-0.50304,0.18194,218.37,0.18775,-0.03963,-0.98142,204.84,0.5009,0.86336,0.060961,263.71,#7,0.8449,-0.50304,0.18194,218.37,0.18775,-0.03963,-0.98142,204.84,0.5009,0.86336,0.060961,263.71
> ui mousemode right "rotate selected models"
> view matrix models
> #3,0.83026,-0.52445,0.18873,218.31,0.038689,-0.28356,-0.95817,204.19,0.55603,0.80284,-0.21514,264.09,#7,0.83026,-0.52445,0.18873,218.31,0.038689,-0.28356,-0.95817,204.19,0.55603,0.80284,-0.21514,264.09
> fitmap #7 inMap #1
Fit molecule fold_gc_diii_36145_fv_model_4.cif (#7) to map
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) using 2549 atoms
average map value = 0.3309, steps = 92
shifted from previous position = 2.38
rotated from previous position = 23.3 degrees
atoms outside contour = 797, contour level = 0.2
Position of fold_gc_diii_36145_fv_model_4.cif (#7) relative to
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) coordinates:
Matrix rotation and translation
0.94449936 -0.21395042 0.24929136 212.07520004
-0.15522514 0.37814154 0.91264129 198.95293941
-0.28952741 -0.90068540 0.32394396 145.72302377
Axis -0.95811497 0.28469792 0.03102892
Axis point 0.00000000 240.54371130 -98.40037650
Rotation angle (degrees) 71.13784566
Shift along axis -142.02930880
> fitmap #3 inMap #1
Fit molecule fold_gc_diii_36145_fv_model_4.cif (#3) to map
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) using 2549 atoms
average map value = 0.3309, steps = 92
shifted from previous position = 2.38
rotated from previous position = 23.3 degrees
atoms outside contour = 797, contour level = 0.2
Position of fold_gc_diii_36145_fv_model_4.cif (#3) relative to
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) coordinates:
Matrix rotation and translation
0.94449936 -0.21395042 0.24929136 212.07520004
-0.15522514 0.37814154 0.91264129 198.95293941
-0.28952741 -0.90068540 0.32394396 145.72302377
Axis -0.95811497 0.28469792 0.03102892
Axis point 0.00000000 240.54371130 -98.40037650
Rotation angle (degrees) 71.13784566
Shift along axis -142.02930880
> select clear
> undo
[Repeated 7 time(s)]
> show #!1 models
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site 6/Models for
> Figure/Gc-42437-36124_refine_035_for domain III.pdb"
Chain information for Gc-42437-36124_refine_035_for domain III.pdb #8
---
Chain | Description
A | No description available
B | No description available
G | No description available
H | No description available
L | No description available
> select add #8
8882 atoms, 9094 bonds, 1 pseudobond, 1154 residues, 2 models selected
> hide (#!8 & sel) target a
> cartoon (#!8 & sel)
> select clear
Drag select of 132 residues
> select up
1350 atoms, 1379 bonds, 177 residues, 1 model selected
> select up
1714 atoms, 1754 bonds, 226 residues, 1 model selected
> cartoon hide sel
Drag select of 128 residues
> select up
1296 atoms, 1322 bonds, 170 residues, 1 model selected
> select up
3361 atoms, 3442 bonds, 443 residues, 1 model selected
> select up
8882 atoms, 9094 bonds, 1154 residues, 1 model selected
> select down
3361 atoms, 3442 bonds, 443 residues, 1 model selected
> cartoon hide sel
Drag select of 353 residues, 1 pseudobonds
> cartoon hide (#!8 & sel)
Drag select of 4 residues
> cartoon hide sel
Drag select of 13 residues
> select clear
Drag select of 10 residues
> select #8/G:1436
10 atoms, 10 bonds, 1 residue, 1 model selected
Drag select of 7 residues
> select up
81 atoms, 80 bonds, 10 residues, 1 model selected
> select up
2155 atoms, 2206 bonds, 276 residues, 1 model selected
> cartoon hide sel
> mmaker #8 to #7
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker fold_gc_diii_36145_fv_model_4.cif, chain A (#7) with
Gc-42437-36124_refine_035_for domain III.pdb, chain G (#8), sequence alignment
score = 520.8
RMSD between 105 pruned atom pairs is 0.570 angstroms; (across all 105 pairs:
0.570)
> select add #8
8882 atoms, 9094 bonds, 1 pseudobond, 1154 residues, 2 models selected
> select subtract #8
Nothing selected
> select add #8
8882 atoms, 9094 bonds, 1 pseudobond, 1154 residues, 2 models selected
> view matrix models
> #8,0.13946,-0.060749,0.98836,211.93,0.54495,-0.82867,-0.12783,275.16,0.82679,0.55644,-0.08246,298.26
> ui mousemode right "translate selected models"
> view matrix models
> #8,0.13946,-0.060749,0.98836,214.69,0.54495,-0.82867,-0.12783,269.87,0.82679,0.55644,-0.08246,298.5
> view matrix models
> #8,0.13946,-0.060749,0.98836,215.21,0.54495,-0.82867,-0.12783,270.32,0.82679,0.55644,-0.08246,295.92
> ui mousemode right "rotate selected models"
> view matrix models
> #8,0.83928,-0.52768,-0.131,264.13,-0.2418,-0.14645,-0.95921,212.98,0.48697,0.83672,-0.25051,271.37
> view matrix models
> #8,0.66366,-0.6478,-0.37404,255.57,-0.31356,0.21306,-0.92536,203.12,0.67914,0.73141,-0.061726,284.42
> view matrix models
> #8,0.51522,-0.709,-0.48151,247.7,-0.082793,0.51802,-0.85135,212.09,0.85305,0.4785,0.2082,298.68
> ui mousemode right "translate selected models"
> view matrix models
> #8,0.51522,-0.709,-0.48151,251.88,-0.082793,0.51802,-0.85135,208.26,0.85305,0.4785,0.2082,289.68
> view matrix models
> #8,0.51522,-0.709,-0.48151,255.82,-0.082793,0.51802,-0.85135,212.59,0.85305,0.4785,0.2082,293.1
> hide #7 models
> hide #3 models
> fitmap #8 inMap #1
Fit molecule Gc-42437-36124_refine_035_for domain III.pdb (#8) to map
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) using 8882 atoms
average map value = 0.176, steps = 272
shifted from previous position = 34
rotated from previous position = 45.5 degrees
atoms outside contour = 5087, contour level = 0.2
Position of Gc-42437-36124_refine_035_for domain III.pdb (#8) relative to
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) coordinates:
Matrix rotation and translation
0.02816358 -0.35713950 -0.93362637 214.16585742
-0.05358637 -0.93319326 0.35535734 192.23128185
-0.99816598 0.04002151 -0.04541987 73.40749618
Axis -0.71273201 0.14587446 0.68610037
Axis point 0.00000000 127.80144058 123.83145931
Rotation angle (degrees) 167.21951974
Shift along axis -74.23631722
> view matrix models
> #8,0.51522,-0.709,-0.48151,279.05,-0.082793,0.51802,-0.85135,231.88,0.85305,0.4785,0.2082,328.9
> select clear
Drag select of 5 residues
> cartoon hide sel
> select add #8
8882 atoms, 9094 bonds, 1 pseudobond, 1154 residues, 2 models selected
> view matrix models
> #8,0.51522,-0.709,-0.48151,259.92,-0.082793,0.51802,-0.85135,213.39,0.85305,0.4785,0.2082,290.21
> fitmap #8 inMap #1
Fit molecule Gc-42437-36124_refine_035_for domain III.pdb (#8) to map
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) using 8882 atoms
average map value = 0.2656, steps = 536
shifted from previous position = 18.6
rotated from previous position = 117 degrees
atoms outside contour = 3344, contour level = 0.2
Position of Gc-42437-36124_refine_035_for domain III.pdb (#8) relative to
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) coordinates:
Matrix rotation and translation
-0.08407712 -0.07018635 0.99398436 188.87403525
-0.45545392 0.88992728 0.02431375 142.98079709
-0.88628030 -0.45066984 -0.10678917 111.57911991
Axis -0.24022687 0.95095933 -0.19485225
Axis point 168.97140736 0.00000000 -22.37136483
Rotation angle (degrees) 98.65413594
Shift along axis 68.85486376
> ui mousemode right "translate selected models"
> ui mousemode right "rotate selected models"
> view matrix models
> #8,0.7769,-0.60798,0.16368,272.92,0.069998,-0.17495,-0.98209,232.7,0.62572,0.77444,-0.09336,272.94
> fitmap #8 inMap #1
Fit molecule Gc-42437-36124_refine_035_for domain III.pdb (#8) to map
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) using 8882 atoms
average map value = 0.07648, steps = 516
shifted from previous position = 27.5
rotated from previous position = 46.4 degrees
atoms outside contour = 7383, contour level = 0.2
Position of Gc-42437-36124_refine_035_for domain III.pdb (#8) relative to
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) coordinates:
Matrix rotation and translation
0.77327299 -0.61398100 0.15835473 249.23314880
-0.53262981 -0.49347488 0.68759583 127.20425412
-0.34402669 -0.61604373 -0.70861538 155.16983898
Axis -0.93153210 0.35898298 0.05813051
Axis point 0.00000000 147.87905583 43.53650009
Rotation angle (degrees) 135.59475538
Shift along axis -177.48441401
> view matrix models
> #8,0.012136,-0.65811,-0.75282,231.06,0.21151,0.73753,-0.64133,226.39,0.9773,-0.15145,0.14815,306.95
> fitmap #8 inMap #1
Fit molecule Gc-42437-36124_refine_035_for domain III.pdb (#8) to map
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) using 8882 atoms
average map value = 0.176, steps = 156
shifted from previous position = 29.9
rotated from previous position = 25.8 degrees
atoms outside contour = 5086, contour level = 0.2
Position of Gc-42437-36124_refine_035_for domain III.pdb (#8) relative to
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) coordinates:
Matrix rotation and translation
0.02822536 -0.35707983 -0.93364732 214.17425912
-0.05317950 -0.93323314 0.35531374 192.26785250
-0.99818599 0.03962204 -0.04533017 73.40133493
Axis -0.71274420 0.14571039 0.68612257
Axis point 0.00000000 127.80819014 123.81373793
Rotation angle (degrees) 167.20507474
Shift along axis -74.27372517
> view matrix models
> #8,0.54929,-0.7777,0.30571,262.85,0.60848,0.1215,-0.78421,258.13,0.57274,0.61678,0.53996,272.4
> view matrix models
> #8,0.98478,0.084306,-0.15198,273.76,-0.16812,0.68367,-0.71016,206.04,0.04403,0.72491,0.68744,241.21
> view matrix models
> #8,0.33928,-0.93695,0.083763,253.62,-0.273,-0.18328,-0.94439,213.69,0.9002,0.29754,-0.31797,295.68
> view matrix models
> #8,0.39462,-0.77374,-0.49557,254.19,0.37213,0.62772,-0.68373,237.06,0.84011,0.085399,0.53565,295.59
> view matrix models
> #8,0.50628,-0.68449,-0.52455,259.04,0.43851,0.7281,-0.52687,239.2,0.74256,0.036726,0.66877,290.9
> ui mousemode right "translate selected models"
> view matrix models
> #8,0.50628,-0.68449,-0.52455,260.85,0.43851,0.7281,-0.52687,232.82,0.74256,0.036726,0.66877,294.4
> view matrix models
> #8,0.50628,-0.68449,-0.52455,259.15,0.43851,0.7281,-0.52687,234.86,0.74256,0.036726,0.66877,292.62
> ui mousemode right "rotate selected models"
> view matrix models
> #8,0.71046,-0.63639,-0.30042,269.79,-0.055177,0.37521,-0.9253,212.82,0.70157,0.67397,0.23146,280.44
> ui mousemode right "translate selected models"
> view matrix models
> #8,0.71046,-0.63639,-0.30042,269.95,-0.055177,0.37521,-0.9253,216.35,0.70157,0.67397,0.23146,280.33
> ui mousemode right "rotate selected models"
> view matrix models
> #8,0.47034,-0.6186,-0.62938,256.28,-0.036628,0.69889,-0.71429,212.34,0.88173,0.35901,0.30606,295.28
> fitmap #8 inMap #1
Fit molecule Gc-42437-36124_refine_035_for domain III.pdb (#8) to map
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) using 8882 atoms
average map value = 0.176, steps = 200
shifted from previous position = 30.5
rotated from previous position = 33.9 degrees
atoms outside contour = 5084, contour level = 0.2
Position of Gc-42437-36124_refine_035_for domain III.pdb (#8) relative to
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) coordinates:
Matrix rotation and translation
0.02846579 -0.35716487 -0.93360749 214.22136234
-0.05361459 -0.93318708 0.35536932 192.20666655
-0.99815589 0.03993912 -0.04571316 73.41212765
Axis -0.71283802 0.14587236 0.68599068
Axis point 0.00000000 127.79484990 123.82971619
Rotation angle (degrees) 167.21756364
Shift along axis -74.30745662
> select subtract #8
Nothing selected
> hide #!8 models
> show #7 models
> show #3 models
> hide #3 models
> select add #7
2549 atoms, 2606 bonds, 331 residues, 1 model selected
> view matrix models
> #7,0.53763,-0.63696,0.55249,220.57,0.58343,-0.19204,-0.78913,242.82,0.60874,0.7466,0.26837,267.73
> ui mousemode right "translate selected models"
> view matrix models
> #7,0.53763,-0.63696,0.55249,220.96,0.58343,-0.19204,-0.78913,206.82,0.60874,0.7466,0.26837,265.4
> ui mousemode right "rotate selected models"
> view matrix models
> #7,0.51686,-0.54473,0.66039,220.92,0.38052,-0.54484,-0.74723,205.9,0.76685,0.63751,-0.074325,265.97
> fitmap #7 inMap #1
Fit molecule fold_gc_diii_36145_fv_model_4.cif (#7) to map
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) using 2549 atoms
average map value = 0.3309, steps = 108
shifted from previous position = 6.67
rotated from previous position = 43.7 degrees
atoms outside contour = 798, contour level = 0.2
Position of fold_gc_diii_36145_fv_model_4.cif (#7) relative to
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) coordinates:
Matrix rotation and translation
0.94450415 -0.21421941 0.24904209 212.07200056
-0.15528555 0.37691342 0.91313891 198.93860506
-0.28947938 -0.90113612 0.32273111 145.69066637
Axis -0.95819556 0.28441602 0.03112547
Axis point 0.00000000 240.33195986 -98.12722032
Rotation angle (degrees) 71.21158121
Shift along axis -142.09043289
> view matrix models
> #7,-0.97139,-0.14226,0.19015,215.31,-0.0019754,-0.79585,-0.60549,203.18,0.23747,-0.58854,0.7728,258.19
> view matrix models
> #7,-0.89834,0.43844,-0.027539,216.64,-0.32528,-0.706,-0.62909,202.85,-0.29526,-0.55618,0.77684,257.4
> fitmap #7 inMap #1
Fit molecule fold_gc_diii_36145_fv_model_4.cif (#7) to map
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) using 2549 atoms
average map value = 0.3013, steps = 84
shifted from previous position = 3.13
rotated from previous position = 30.9 degrees
atoms outside contour = 885, contour level = 0.2
Position of fold_gc_diii_36145_fv_model_4.cif (#7) relative to
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) coordinates:
Matrix rotation and translation
-0.83487440 0.22876868 -0.50064921 210.74752356
0.06868658 0.94573180 0.31760591 199.58572652
0.54613816 0.23077316 -0.80528061 152.65225563
Axis -0.08172408 -0.98520128 -0.15066392
Axis point 80.02337574 0.00000000 89.00037741
Rotation angle (degrees) 147.90967095
Shift along axis -236.85444724
> view matrix models
> #7,0.48493,0.48601,-0.72707,219.45,-0.13988,-0.77756,-0.61305,204.64,-0.86329,0.399,-0.30908,257.39
> fitmap #7 inMap #1
Fit molecule fold_gc_diii_36145_fv_model_4.cif (#7) to map
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) using 2549 atoms
average map value = 0.3309, steps = 168
shifted from previous position = 3.89
rotated from previous position = 78.8 degrees
atoms outside contour = 798, contour level = 0.2
Position of fold_gc_diii_36145_fv_model_4.cif (#7) relative to
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) coordinates:
Matrix rotation and translation
0.94451810 -0.21410467 0.24908787 212.07272100
-0.15522013 0.37737858 0.91295790 198.93861830
-0.28946897 -0.90096870 0.32320755 145.69455183
Axis -0.95817618 0.28448358 0.03110477
Axis point 0.00000000 240.40110891 -98.22814510
Rotation angle (degrees) 71.18266373
Shift along axis -142.07646398
> select subtract #7
Nothing selected
> show #3 models
> hide #3 models
> show #!8 models
> select add #8
8882 atoms, 9094 bonds, 1 pseudobond, 1154 residues, 2 models selected
> view matrix models
> #8,0.7059,-0.56105,-0.43235,268.53,-0.30091,0.31504,-0.90011,203.57,0.64122,0.76549,0.053564,275.55
> view matrix models
> #8,0.7488,-0.47118,-0.46615,269.52,-0.30705,0.37668,-0.87397,202.27,0.58739,0.79756,0.13739,272.04
> fitmap #8 inMap #1
Fit molecule Gc-42437-36124_refine_035_for domain III.pdb (#8) to map
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) using 8882 atoms
average map value = 0.116, steps = 216
shifted from previous position = 25.8
rotated from previous position = 44.7 degrees
atoms outside contour = 6444, contour level = 0.2
Position of Gc-42437-36124_refine_035_for domain III.pdb (#8) relative to
cryosparc_P409_J122_004_volume_map_sharp.mrc (#1) coordinates:
Matrix rotation and translation
0.62236525 0.14584208 -0.76901989 252.45418862
0.72587766 -0.47513329 0.49734292 230.82561825
-0.29285342 -0.86774330 -0.40156996 157.09268783
Axis -0.87630997 -0.30567257 0.37235080
Axis point 0.00000000 138.01238368 88.61468366
Rotation angle (degrees) 128.84156430
Shift along axis -233.29159342
> view matrix models
> #8,0.68087,-0.3793,-0.62654,264.31,-0.43381,0.48039,-0.76225,193.57,0.59011,0.79079,0.16254,272.3
> select subtract #8
Nothing selected
> hide #!8 models
> volume #1 level 0.25
> volume #1 level 0.22
> volume #1 level 0.2
> volume #1 level 0.22
> volume #1 level 0.2
> hide #!1 models
> hide #!4 models
> hide #!5 models
> show #!4 models
> hide #!4 models
> hide #7 models
> show #7 models
> hide #7 models
> turn y 90
[Repeated 3 time(s)]
> view matrix
view matrix camera
0.36402,0.091799,-0.92685,-110.64,0.85547,-0.42649,0.29374,308.03,-0.36833,-0.89982,-0.23378,121.91
view matrix models
#1,0.99758,-0.031402,-0.061997,21.992,-0.025974,-0.99591,0.086502,395.32,-0.06446,-0.084682,-0.99432,437.53,#1.1,1,0,0,0,0,1,0,0,0,0,1,0,#2,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#2.1,1,0,0,0,0,1,0,0,0,0,1,0,#3,-0.51773,0.73064,0.4451,221.21,0.54891,-0.11538,0.82788,240.6,0.65624,0.67294,-0.34132,268.55,#4,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#4.1,1,0,0,0,0,1,0,0,0,0,1,0,#5,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#5.1,1,0,0,0,0,1,0,0,0,0,1,0,#6,-0.014521,-0.06976,0.99746,199.01,0.37884,-0.92358,-0.059077,257.64,0.92535,0.37702,0.039838,302.31,#6.1,1,0,0,0,0,1,0,0,0,0,1,0,#7,0.96505,-0.16958,0.19978,218.27,0.10501,-0.44821,-0.88774,204.29,0.24009,0.8777,-0.41474,262.15,#8,0.68087,-0.3793,-0.62654,264.31,-0.43381,0.48039,-0.76225,193.57,0.59011,0.79079,0.16254,272.3,#8.1,1,0,0,0,0,1,0,0,0,0,1,0
> save C:\Users\ckh755\Desktop\image1.png supersample 3
> dssp
> turn y 90
[Repeated 3 time(s)]
> view matrix
view matrix camera
0.39622,0.05884,-0.91627,-108.65,0.84363,-0.41716,0.33802,323.39,-0.36234,-0.90693,-0.21493,127.92
view matrix models
#1,0.99758,-0.031402,-0.061997,21.992,-0.025974,-0.99591,0.086502,395.32,-0.06446,-0.084682,-0.99432,437.53,#1.1,1,0,0,0,0,1,0,0,0,0,1,0,#2,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#2.1,1,0,0,0,0,1,0,0,0,0,1,0,#3,-0.51773,0.73064,0.4451,221.21,0.54891,-0.11538,0.82788,240.6,0.65624,0.67294,-0.34132,268.55,#4,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#4.1,1,0,0,0,0,1,0,0,0,0,1,0,#5,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#5.1,1,0,0,0,0,1,0,0,0,0,1,0,#6,-0.014521,-0.06976,0.99746,199.01,0.37884,-0.92358,-0.059077,257.64,0.92535,0.37702,0.039838,302.31,#6.1,1,0,0,0,0,1,0,0,0,0,1,0,#7,0.96505,-0.16958,0.19978,218.27,0.10501,-0.44821,-0.88774,204.29,0.24009,0.8777,-0.41474,262.15,#8,0.68087,-0.3793,-0.62654,264.31,-0.43381,0.48039,-0.76225,193.57,0.59011,0.79079,0.16254,272.3,#8.1,1,0,0,0,0,1,0,0,0,0,1,0
> save C:\Users\ckh755\Desktop\image2.png supersample 3
> show #!1 models
> show #3 models
> hide #3 models
> show #!4 models
> show #!5 models
> show #7 models
> show #!8 models
> hide #!8 models
> show #3 models
> mmaker #3 to #7
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker fold_gc_diii_36145_fv_model_4.cif, chain B (#7) with
fold_gc_diii_36145_fv_model_4.cif, chain B (#3), sequence alignment score =
618.1
RMSD between 118 pruned atom pairs is 0.000 angstroms; (across all 118 pairs:
0.000)
> hide #7 models
> hide #3 models
> show #3 models
> close #7#8
> hide #!5 models
> hide #!4 models
> hide #3 models
> hide #!2 models
> hide #!1 models
> select #6/A:108-214
826 atoms, 842 bonds, 107 residues, 1 model selected
> hide sel
> select #6/B:114-220
771 atoms, 789 bonds, 107 residues, 1 model selected
> hide sel
> select #6/A:108-214
826 atoms, 842 bonds, 107 residues, 1 model selected
> hide cartoons sel
Expected ',' or a keyword
> select #6/B:114-220
771 atoms, 789 bonds, 107 residues, 1 model selected
> hide cartoons sel
Expected ',' or a keyword
> hide sel cartoons
> undo
> select #6/A:108-214
826 atoms, 842 bonds, 107 residues, 1 model selected
> hide sel cartoons
> select #6/B:114-220
771 atoms, 789 bonds, 107 residues, 1 model selected
> hide sel cartoons
> show #!5 models
> show #!4 models
> show #3 models
> show #!2 models
> show #!1 models
> ui tool show "Map Eraser"
> volume erase #1 center 167.41,169.4,228.53 radius 16.921
> hide #7 models
> close #7
> select add #6
8882 atoms, 9094 bonds, 1 pseudobond, 1154 residues, 2 models selected
> select subtract #6
Nothing selected
> save "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Figure #_Site 6/8-15-2024_Site 6 models and J122 map for
> figure.cxs" includeMaps true
——— End of log from Thu Aug 15 18:02:26 2024 ———
opened ChimeraX session
> hide #!1 models
> hide #!2 models
> show #!2 models
> hide #3 models
> show #3 models
> select add #4
5978 atoms, 6127 bonds, 4 pseudobonds, 772 residues, 2 models selected
> select add #5
11956 atoms, 12254 bonds, 8 pseudobonds, 1544 residues, 4 models selected
> select add #6
20838 atoms, 21348 bonds, 9 pseudobonds, 2698 residues, 6 models selected
> lighting soft
> dssp
> graphics selection color black
> select add #3
23387 atoms, 23954 bonds, 9 pseudobonds, 3029 residues, 7 models selected
> select add #2
29365 atoms, 30081 bonds, 13 pseudobonds, 3801 residues, 9 models selected
> graphics selection color black
> select #2-6
29365 atoms, 30081 bonds, 13 pseudobonds, 3801 residues, 9 models selected
> select subtract #6
20483 atoms, 20987 bonds, 12 pseudobonds, 2647 residues, 7 models selected
> select add #6
29365 atoms, 30081 bonds, 13 pseudobonds, 3801 residues, 9 models selected
> select subtract #5
23387 atoms, 23954 bonds, 9 pseudobonds, 3029 residues, 7 models selected
> select clear
Drag select of 43 residues
> select clear
> select add #6
8882 atoms, 9094 bonds, 1 pseudobond, 1154 residues, 2 models selected
> select add #5
14860 atoms, 15221 bonds, 5 pseudobonds, 1926 residues, 4 models selected
> select add #4
20838 atoms, 21348 bonds, 9 pseudobonds, 2698 residues, 6 models selected
> select add #2
26816 atoms, 27475 bonds, 13 pseudobonds, 3470 residues, 8 models selected
> select subtract #2
20838 atoms, 21348 bonds, 9 pseudobonds, 2698 residues, 6 models selected
> select add #2
26816 atoms, 27475 bonds, 13 pseudobonds, 3470 residues, 8 models selected
> select subtract #2
20838 atoms, 21348 bonds, 9 pseudobonds, 2698 residues, 6 models selected
> select subtract #4
14860 atoms, 15221 bonds, 5 pseudobonds, 1926 residues, 4 models selected
> select subtract #5
8882 atoms, 9094 bonds, 1 pseudobond, 1154 residues, 2 models selected
> select subtract #6
Nothing selected
Drag select of 48 residues
> select up
978 atoms, 998 bonds, 125 residues, 1 model selected
> select up
1714 atoms, 1756 bonds, 226 residues, 1 model selected
> select up
5978 atoms, 6127 bonds, 772 residues, 1 model selected
> select up
29365 atoms, 30081 bonds, 3801 residues, 7 models selected
> select down
5978 atoms, 6127 bonds, 772 residues, 1 model selected
> graphics selection color green
> graphics selection color limegreen
> select clear
Drag select of 43 residues
> select up
503 atoms, 512 bonds, 68 residues, 1 model selected
> select up
1714 atoms, 1756 bonds, 226 residues, 1 model selected
> surface sel
Drag select of 25 residues
> select up
699 atoms, 708 bonds, 86 residues, 1 model selected
> select up
3361 atoms, 3442 bonds, 443 residues, 1 model selected
> surface sel
Drag select of 54 residues
> select up
747 atoms, 758 bonds, 98 residues, 1 model selected
> select up
1738 atoms, 1784 bonds, 225 residues, 1 model selected
> select up
5978 atoms, 6127 bonds, 772 residues, 1 model selected
> select up
29365 atoms, 30081 bonds, 3801 residues, 7 models selected
> select down
5978 atoms, 6127 bonds, 772 residues, 5 models selected
> surface sel
> undo
[Repeated 1 time(s)]
> surface hidePatches (#!5 & sel)
> select down
1738 atoms, 1784 bonds, 225 residues, 6 models selected
> select down
747 atoms, 758 bonds, 98 residues, 3 models selected
> select up
1738 atoms, 1784 bonds, 225 residues, 3 models selected
> surface (#!5 & sel)
Drag select of 57 residues
> select up
922 atoms, 943 bonds, 119 residues, 1 model selected
> select up
1753 atoms, 1794 bonds, 226 residues, 1 model selected
> surface sel
> select clear
Drag select of Gc-42437-36124_refine_035_for 36124 Fab model.pdb_A SES
surface, 29026 of 192580 triangles, Gc-42437-36124_refine_035_for 36124 Fab
model.pdb_B SES surface, 16942 of 200702 triangles, 2 residues
> select up
80 atoms, 80 bonds, 12 residues, 3 models selected
> select up
3361 atoms, 3442 bonds, 443 residues, 3 models selected
> surface hidePatches (#!6 & sel)
> select clear
Drag select of 97 residues
> select up
1156 atoms, 1179 bonds, 145 residues, 3 models selected
> select up
3361 atoms, 3442 bonds, 443 residues, 3 models selected
> surface (#!6 & sel)
> hide #!6 models
> show #!6 models
> select add #6
8882 atoms, 9094 bonds, 1 pseudobond, 1154 residues, 4 models selected
> select subtract #6
2 models selected
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Structure Files/Gc-4Ffabs/Map for Gc245 med res map with Site 6 and
> domain III/fold_gc_diii_36145_fv_model_4.cif"
Chain information for fold_gc_diii_36145_fv_model_4.cif #7
---
Chain | Description
A | .
B | .
C | .
> mm#7 to #3
Unknown command: mm#7 to #3
> mmaker #7 to #3
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker fold_gc_diii_36145_fv_model_4.cif, chain B (#3) with
fold_gc_diii_36145_fv_model_4.cif, chain B (#7), sequence alignment score =
618.1
RMSD between 118 pruned atom pairs is 0.000 angstroms; (across all 118 pairs:
0.000)
> hide #!6 models
> hide #!5 models
> hide #!4 models
> hide #!3 models
> hide #!2 models
> hide #7 models
> show #7 models
Drag select of 34 residues
> select up
366 atoms, 371 bonds, 47 residues, 1 model selected
> select up
796 atoms, 808 bonds, 105 residues, 1 model selected
> delete atoms sel
> delete bonds sel
> close #7
> show #!6 models
> show #!5 models
> show #!4 models
> show #!3 models
> show #!2 models
> hide #!5 models
> hide #!4 models
> hide #!3 models
> hide #!2 models
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Structure Files/Gc-4Ffabs/Building for Gc-2-4-5 str/Combined
> Models/AF2_37847_e61a7.result/37847_e61a7/37847_e61a7_unrelaxed_rank_001_alphafold2_multimer_v3_model_1_seed_000.pdb"
Chain information for
37847_e61a7_unrelaxed_rank_001_alphafold2_multimer_v3_model_1_seed_000.pdb #7
---
Chain | Description
A | No description available
B | No description available
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Structure Files/Gc-4Ffabs/Building for Gc-2-4-5 str/Combined
> Models/AF2_36145_203f6.result/36145_203f6/36145_203f6_unrelaxed_rank_001_alphafold2_multimer_v3_model_4_seed_000.pdb"
Chain information for
36145_203f6_unrelaxed_rank_001_alphafold2_multimer_v3_model_4_seed_000.pdb #8
---
Chain | Description
A | No description available
B | No description available
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Structure Files/Gc-4Ffabs/Building for Gc-2-4-5 str/Combined
> Models/Gc-dI-dII_42437_37847_36124.pdb"
Chain information for Gc-dI-dII_42437_37847_36124.pdb #9
---
Chain | Description
A | No description available
B | No description available
C | No description available
D | No description available
G | No description available
H | No description available
L | No description available
> hide #7 models
> hide #8 models
> hide #!6 models
Drag select of 7 atoms
> delete atoms sel
> delete bonds sel
> hide #!9 models
> show #7 models
> mmaker #7 to #5
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for 37847
Fv model.pdb, chain A (#5) with
37847_e61a7_unrelaxed_rank_001_alphafold2_multimer_v3_model_1_seed_000.pdb,
chain A (#7), sequence alignment score = 562.8
RMSD between 116 pruned atom pairs is 0.513 angstroms; (across all 116 pairs:
0.513)
> show #!5 models
> hide #!5 models
> show #8 models
> hide #7 models
> show #!3 models
> mmaker #8 to #3
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker fold_gc_diii_36145_fv_model_4.cif, chain B (#3) with
36145_203f6_unrelaxed_rank_001_alphafold2_multimer_v3_model_4_seed_000.pdb,
chain A (#8), sequence alignment score = 614.5
RMSD between 118 pruned atom pairs is 0.257 angstroms; (across all 118 pairs:
0.257)
> show #!9 models
> hide #!3 models
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Structure Files/Gc-4Ffabs/Building for Gc-2-4-5 str/Combined
> Models/Gc-dI-dII_42437_37847_36124.pdb"
Chain information for Gc-dI-dII_42437_37847_36124.pdb #10
---
Chain | Description
A | No description available
B | No description available
C | No description available
D | No description available
G | No description available
H | No description available
L | No description available
Drag select of 7 atoms
> delete atoms sel
> delete bonds sel
> hide #8 models
Drag select of 13564 atoms, 4 pseudobonds, 84 bonds
> select up
13864 atoms, 14186 bonds, 4 pseudobonds, 1786 residues, 4 models selected
> select up
15188 atoms, 15556 bonds, 4 pseudobonds, 1964 residues, 4 models selected
> delete atoms (#!9-10 & sel)
> delete bonds (#!9-10 & sel)
Drag select of 290 atoms, 2 pseudobonds
> select clear
Drag select of 394 atoms, 6 pseudobonds
> delete atoms (#!9-10 & sel)
> delete bonds (#!9-10 & sel)
Drag select of 190 atoms, 4 pseudobonds
> select up
206 atoms, 200 bonds, 4 pseudobonds, 24 residues, 4 models selected
> select up
350 atoms, 342 bonds, 4 pseudobonds, 46 residues, 4 models selected
> select up
458 atoms, 456 bonds, 4 pseudobonds, 60 residues, 4 models selected
> select up
586 atoms, 584 bonds, 4 pseudobonds, 76 residues, 4 models selected
> select up
7328 atoms, 7486 bonds, 8 pseudobonds, 964 residues, 4 models selected
> select down
586 atoms, 584 bonds, 4 pseudobonds, 76 residues, 4 models selected
> delete atoms (#!9-10 & sel)
> delete bonds (#!9-10 & sel)
> hide #!9 models
> hide #!10 models
> show #!2 models
> show #!3 models
> show #8 models
> color #8/B,C #DD833E
> color #8 #DD833E
> hide #!3 models
> hide #8 models
> show #!3 models
Drag select of fold_gc_diii_36145_fv_model_4.cif_B SES surface, 37504 of
104344 triangles, fold_gc_diii_36145_fv_model_4.cif_C SES surface, 30389 of
92906 triangles, 107 residues
> select up
1216 atoms, 1241 bonds, 157 residues, 3 models selected
> select up
1753 atoms, 1794 bonds, 226 residues, 3 models selected
> surface hidePatches (#!3 & sel)
> cartoon hide (#!3 & sel)
> select add #3
2549 atoms, 2606 bonds, 331 residues, 3 models selected
> select subtract #3
2 models selected
> show #8 models
> select add #8
3340 atoms, 3417 bonds, 439 residues, 1 model selected
> surface sel
> select subtract #8
2 models selected
> hide #!8 models
> hide #!3 models
> hide #!2 models
> show #!4 models
> hide #!4 models
> show #!4 models
> show #!9 models
> mmaker #9 to #4
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for 42437
Fv model.pdb, chain B (#4) with Gc-dI-dII_42437_37847_36124.pdb, chain C (#9),
sequence alignment score = 478.5
RMSD between 99 pruned atom pairs is 0.674 angstroms; (across all 108 pairs:
1.655)
> mmaker #9 to #4/H
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker
Gc-42437-36124_37847_refined_041-Isolde_real_space_refined_350-noH_for 42437
Fv model.pdb, chain H (#4) with Gc-dI-dII_42437_37847_36124.pdb, chain D (#9),
sequence alignment score = 334.5
RMSD between 106 pruned atom pairs is 0.765 angstroms; (across all 117 pairs:
1.510)
> hide #!4 models
> show #!4 models
> hide #!4 models
> select add #9
3371 atoms, 3451 bonds, 444 residues, 1 model selected
> hide sel target a
> surface sel
> cartoon (#!9 & sel)
> color #9 #9142AE
> select subtract #9
2 models selected
> hide #!9 models
> show #!10 models
> color #10 #CC66A7
> hide #10 target a
> cartoon #10
> surface #10
> show #!6 models
> hide #!10 models
> hide #!6 models
> show #!2 models
> show #!3 models
> show #!4 models
> show #!5 models
> show #!6 models
> show #7 models
> hide #!5 models
> select add #7
3403 atoms, 3484 bonds, 448 residues, 1 model selected
> surface sel
> select subtract #7
2 models selected
> hide #!7 models
> show #!7 models
> show #!5 models
> hide #!5 models
> show #!8 models
> show #!9 models
> hide #!4 models
> show #!10 models
> close #10
> show #!4 models
> hide #!4 models
> show #!5 models
> hide #!5 models
> hide #!9 models
> show #!9 models
> hide #!9 models
> show #!9 models
> rename #9 "42437 xstal model"
> rename #9 "42437 Fab xstal model"
> hide #!8 models
> show #!8 models
> rename #8 "AF2 36145 Fab"
> hide #!7 models
> show #!7 models
> rename #7 "AF2 37847 Fab"
> hide #!6 models
> show #!6 models
> rename #6 "36124 Fab crystal"
> show #!5 models
> hide #!5 models
> show #!5 models
> hide #!5 models
> close #4-5
> hide #!3 models
> show #!3 models
> rename #3 "AF3_domain III from bound w36145Fv"
> rename #2 "Gc DI-DII from Gc/2/5xstal"
> hide #!2 models
> show #!2 models
> hide #!3 models
> show #!3 models
> hide #!6 models
> show #!6 models
> hide #!7 models
> show #!7 models
> hide #!8 models
> show #!8 models
> hide #!9 models
> show #!9 models
> transparency #2-3,6-9 50
> transparency #2-3,6-9 20
> transparency #2-3,6-9 30
> select add #6
8882 atoms, 9094 bonds, 1 pseudobond, 1154 residues, 2 models selected
> cartoon (#!6 & sel)
> undo
> hide #!3 models
> hide #!2 models
> hide #!7 models
> hide #!8 models
> hide #!9 models
> select #6/A:202@CB
1 atom, 1 residue, 1 model selected
Drag select of Gc-42437-36124_refine_035_for 36124 Fab model.pdb_A SES
surface, 137335 of 192580 triangles, Gc-42437-36124_refine_035_for 36124 Fab
model.pdb_B SES surface, 147969 of 200702 triangles, 160 residues
> select up
1502 atoms, 1534 bonds, 193 residues, 3 models selected
> select up
3361 atoms, 3442 bonds, 443 residues, 3 models selected
> cartoon (#!6 & sel)
> show #!3 models
> show #!2 models
> show #!7 models
> show #!8 models
> show #!9 models
> select add #6
8882 atoms, 9094 bonds, 1 pseudobond, 1154 residues, 4 models selected
> select subtract #6
2 models selected
> select clear
> transparency #2-3,6-9 80
> transparency #2-3,6-9 90
> transparency #2-3,6-9 60
> select clear
> view matrix camera
> -0.056974,-0.13241,-0.98956,-198.4,0.90561,-0.42408,0.0045992,209.03,-0.42025,-0.8959,0.14407,268.38
> view matrix models
> #1,0.99758,-0.031402,-0.061997,21.992,-0.025974,-0.99591,0.086502,395.32,-0.06446,-0.084682,-0.99432,437.53,#1.1,1,0,0,0,0,1,0,0,0,0,1,0,#2,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#2.1,1,0,0,0,0,1,0,0,0,0,1,0,#3,0.96505,-0.16958,0.19978,218.27,0.10501,-0.44821,-0.88774,204.29,0.24009,0.8777,-0.41474,262.15,#4,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#4.1,1,0,0,0,0,1,0,0,0,0,1,0,#5,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#5.1,1,0,0,0,0,1,0,0,0,0,1,0,#6,-0.014521,-0.06976,0.99746,199.01,0.37884,-0.92358,-0.059077,257.64,0.92535,0.37702,0.039838,302.31,#6.1,1,0,0,0,0,1,0,0,0,0,1,0
Invalid "models" argument: No models specified by "#4"
> view matrix camera
> 0.39622,0.05884,-0.91627,-108.65,0.84363,-0.41716,0.33802,323.39,-0.36234,-0.90693,-0.21493,127.92
> view matrix models
> #1,0.99758,-0.031402,-0.061997,21.992,-0.025974,-0.99591,0.086502,395.32,-0.06446,-0.084682,-0.99432,437.53,#1.1,1,0,0,0,0,1,0,0,0,0,1,0,#2,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#2.1,1,0,0,0,0,1,0,0,0,0,1,0,#3,-0.51773,0.73064,0.4451,221.21,0.54891,-0.11538,0.82788,240.6,0.65624,0.67294,-0.34132,268.55,#4,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#4.1,1,0,0,0,0,1,0,0,0,0,1,0,#5,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#5.1,1,0,0,0,0,1,0,0,0,0,1,0,#6,-0.014521,-0.06976,0.99746,199.01,0.37884,-0.92358,-0.059077,257.64,0.92535,0.37702,0.039838,302.31,#6.1,1,0,0,0,0,1,0,0,0,0,1,0,#7,0.96505,-0.16958,0.19978,218.27,0.10501,-0.44821,-0.88774,204.29,0.24009,0.8777,-0.41474,262.15,#8,0.68087,-0.3793,-0.62654,264.31,-0.43381,0.48039,-0.76225,193.57,0.59011,0.79079,0.16254,272.3,#8.1,1,0,0,0,0,1,0,0,0,0,1,0
Invalid "models" argument: No models specified by "#4"
> view matrix camera
> -0.056974,-0.13241,-0.98956,-198.4,0.90561,-0.42408,0.0045992,209.03,-0.42025,-0.8959,0.14407,268.38
> view matrix models
> #1,0.99758,-0.031402,-0.061997,21.992,-0.025974,-0.99591,0.086502,395.32,-0.06446,-0.084682,-0.99432,437.53,#1.1,1,0,0,0,0,1,0,0,0,0,1,0,#2,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#2.1,1,0,0,0,0,1,0,0,0,0,1,0,#3,0.96505,-0.16958,0.19978,218.27,0.10501,-0.44821,-0.88774,204.29,0.24009,0.8777,-0.41474,262.15,#4,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#4.1,1,0,0,0,0,1,0,0,0,0,1,0,#5,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#5.1,1,0,0,0,0,1,0,0,0,0,1,0,#6,-0.014521,-0.06976,0.99746,199.01,0.37884,-0.92358,-0.059077,257.64,0.92535,0.37702,0.039838,302.31,#6.1,1,0,0,0,0,1,0,0,0,0,1,0
Invalid "models" argument: No models specified by "#4"
> view matrix camera
> -0.056974,-0.13241,-0.98956,-198.4,0.90561,-0.42408,0.0045992,209.03,-0.42025,-0.8959,0.14407,268.38
> view matrix models
> #1,0.99758,-0.031402,-0.061997,21.992,-0.025974,-0.99591,0.086502,395.32,-0.06446,-0.084682,-0.99432,437.53,#1.1,1,0,0,0,0,1,0,0,0,0,1,0,#2,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#2.1,1,0,0,0,0,1,0,0,0,0,1,0,#3,0.96505,-0.16958,0.19978,218.27,0.10501,-0.44821,-0.88774,204.29,0.24009,0.8777,-0.41474,262.15,#4,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#4.1,1,0,0,0,0,1,0,0,0,0,1,0,#5,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#5.1,1,0,0,0,0,1,0,0,0,0,1,0,#6,-0.014521,-0.06976,0.99746,199.01,0.37884,-0.92358,-0.059077,257.64,0.92535,0.37702,0.039838,302.31,#6.1,1,0,0,0,0,1,0,0,0,0,1,0
Invalid "models" argument: No models specified by "#4"
> color #7 #CC66A7
> transparency #2-3,6-9 60
> save 9-4-2024-Gc4Fabs-modes_1.png transparentBackground true
> transparency #2-3,6-9 50
> transparency #2-3,6-9 40
> save 9-4-2024-Gc4Fabs-modes_2.png transparentBackground true
> open 7a59
7a59 title:
Crimean-Congo Hemorrhagic Fever Virus Envelope Glycoprotein Gc
W1191H/W1197A/W1199A Mutant in Postfusion Conformation (Orthorhombic Crystal
Form) [more info...]
Chain information for 7a59 #4
---
Chain | Description | UniProt
A B C | Envelopment polyprotein | GP_CCHFI 1041-1561
Non-standard residues in 7a59 #4
---
BMA — beta-D-mannopyranose (beta-D-mannose; D-mannose; mannose)
CL — chloride ion
FUC — alpha-L-fucopyranose (alpha-L-fucose; 6-deoxy-alpha-L-galactopyranose;
L-fucose; fucose)
MAN — alpha-D-mannopyranose (alpha-D-mannose; D-mannose; mannose)
NAG — 2-acetamido-2-deoxy-beta-D-glucopyranose (N-acetyl-beta-D-glucosamine;
2-acetamido-2-deoxy-beta-D-glucose; 2-acetamido-2-deoxy-D-glucose;
2-acetamido-2-deoxy-glucose; N-ACETYL-D-GLUCOSAMINE)
PO4 — phosphate ion
> close #4
> close #1
> open 7a59
7a59 title:
Crimean-Congo Hemorrhagic Fever Virus Envelope Glycoprotein Gc
W1191H/W1197A/W1199A Mutant in Postfusion Conformation (Orthorhombic Crystal
Form) [more info...]
Chain information for 7a59 #1
---
Chain | Description | UniProt
A B C | Envelopment polyprotein | GP_CCHFI 1041-1561
Non-standard residues in 7a59 #1
---
BMA — beta-D-mannopyranose (beta-D-mannose; D-mannose; mannose)
CL — chloride ion
FUC — alpha-L-fucopyranose (alpha-L-fucose; 6-deoxy-alpha-L-galactopyranose;
L-fucose; fucose)
MAN — alpha-D-mannopyranose (alpha-D-mannose; D-mannose; mannose)
NAG — 2-acetamido-2-deoxy-beta-D-glucopyranose (N-acetyl-beta-D-glucosamine;
2-acetamido-2-deoxy-beta-D-glucose; 2-acetamido-2-deoxy-D-glucose;
2-acetamido-2-deoxy-glucose; N-ACETYL-D-GLUCOSAMINE)
PO4 — phosphate ion
> select #1/A: 1041-1140, 1232-1268, 1419-1430
954 atoms, 965 bonds, 1 pseudobond, 126 residues, 2 models selected
> color sel #FF7F7F
> select #1/A: 1141-1186, 1205-1231, 1269-1282, 1296-1320, 1347-1403,
> 1410-1418, 1187-1204
1548 atoms, 1595 bonds, 196 residues, 1 model selected
> color sel #FFFF7F
> select #1/A: 1283-1295, 1321-1346, 1404-1409
364 atoms, 374 bonds, 45 residues, 1 model selected
> color sel #c18c7f
> select #1/A: 1431-1441
90 atoms, 90 bonds, 11 residues, 1 model selected
> color sel #7FFFFF
> select #1/A: 1442-1546
795 atoms, 811 bonds, 105 residues, 1 model selected
> color sel #7F7FFF
> select #1/A: 1547-1572
123 atoms, 124 bonds, 16 residues, 1 model selected
> color sel #FF7FFF
> cartoon hide #1/B
> surface #1/B
> color #1/B dark gray
> transparency #1/B 30
> cartoon hide #1/C
> surface #1/C
> color #1/C white
> transparency #1/C 30
> save "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Supplemental Figure #_modes of neut/9-4-2024_Gc with all 4
> Sites models.cxs"
> mmaker #1/A to #2
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker Gc DI-DII from Gc/2/5xstal, chain G (#2) with 7a59, chain A (#1),
sequence alignment score = 1456.8
RMSD between 260 pruned atom pairs is 0.937 angstroms; (across all 321 pairs:
3.709)
> hide #!2 models
> hide #!3 models
> hide #!6 models
> hide #!7 models
> hide #!8 models
> hide #!9 models
Drag select of 7a59_B SES surface, 206050 of 450426 triangles, 7a59_C SES
surface, 189804 of 434220 triangles, 216 atoms, 231 residues, 208 bonds
> hide (#!1 & sel) target a
> show #1/B cartoons
> color #1/B dark gray
> show #1/C cartoons
> color #1/C white
> surface hidePatches (#!1 & sel)
[Repeated 1 time(s)]
> select #1
12589 atoms, 12161 bonds, 3 pseudobonds, 2256 residues, 2 models selected
> surface hidePatches (#!1 & sel)
> select clear
> show #!2 models
> hide #!2 models
> show #!6 models
> show #!7 models
> show #!8 models
> show #!9 models
> view matrix camera
> -0.056974,-0.13241,-0.98956,-198.4,0.90561,-0.42408,0.0045992,209.03,-0.42025,-0.8959,0.14407,268.38
> view matrix models
> #1,0.99758,-0.031402,-0.061997,21.992,-0.025974,-0.99591,0.086502,395.32,-0.06446,-0.084682,-0.99432,437.53,#1.1,1,0,0,0,0,1,0,0,0,0,1,0,#2,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#2.1,1,0,0,0,0,1,0,0,0,0,1,0,#3,0.96505,-0.16958,0.19978,218.27,0.10501,-0.44821,-0.88774,204.29,0.24009,0.8777,-0.41474,262.15,#4,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#4.1,1,0,0,0,0,1,0,0,0,0,1,0,#5,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#5.1,1,0,0,0,0,1,0,0,0,0,1,0,#6,-0.014521,-0.06976,0.99746,199.01,0.37884,-0.92358,-0.059077,257.64,0.92535,0.37702,0.039838,302.31,#6.1,1,0,0,0,0,1,0,0,0,0,1,0
Invalid "models" argument: No models specified by "#4"
> turn y 180
> view matrix
view matrix camera
0.056975,-0.13241,0.98956,646.51,-0.90561,-0.42408,-0.0046034,248.75,0.42026,-0.8959,-0.14408,124.7
view matrix models
#2,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#2.1,1,0,0,0,0,1,0,0,0,0,1,0,#3,0.96505,-0.16958,0.19978,218.27,0.10501,-0.44821,-0.88774,204.29,0.24009,0.8777,-0.41474,262.15,#3.1,1,0,0,0,0,1,0,0,0,0,1,0,#3.2,1,0,0,0,0,1,0,0,0,0,1,0,#6,-0.014521,-0.06976,0.99746,199.01,0.37884,-0.92358,-0.059077,257.64,0.92535,0.37702,0.039838,302.31,#6.1,1,0,0,0,0,1,0,0,0,0,1,0,#6.2,1,0,0,0,0,1,0,0,0,0,1,0,#6.3,1,0,0,0,0,1,0,0,0,0,1,0,#7,0.8169,-0.21413,0.53556,204.47,0.42791,-0.39761,-0.81167,168.64,0.38675,0.89222,-0.23318,214.6,#7.1,1,0,0,0,0,1,0,0,0,0,1,0,#7.2,1,0,0,0,0,1,0,0,0,0,1,0,#8,0.99431,0.074735,-0.075973,218.33,-0.026887,0.86575,0.49976,177.43,0.10312,-0.49487,0.86283,264.22,#8.1,1,0,0,0,0,1,0,0,0,0,1,0,#8.2,1,0,0,0,0,1,0,0,0,0,1,0,#9,0.296,-0.067032,0.95283,-8.4654,0.64478,-0.72195,-0.25109,250.56,0.70473,0.68869,-0.17048,-85.496,#9.1,1,0,0,0,0,1,0,0,0,0,1,0,#9.2,1,0,0,0,0,1,0,0,0,0,1,0,#9.3,1,0,0,0,0,1,0,0,0,0,1,0,#1,-0.9757,0.21626,0.035289,201.77,-0.14806,-0.76939,0.62138,181.78,0.16153,0.60106,0.78271,185.59,#1.1,1,0,0,0,0,1,0,0,0,0,1,0,#1.2,1,0,0,0,0,1,0,0,0,0,1,0,#1.3,1,0,0,0,0,1,0,0,0,0,1,0
> show #!2 models
> show #!3 models
> view matrix
view matrix camera
0.056975,-0.13241,0.98956,646.95,-0.90561,-0.42408,-0.0046034,204.41,0.42026,-0.8959,-0.14408,129.14
view matrix models
#2,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#2.1,1,0,0,0,0,1,0,0,0,0,1,0,#3,0.96505,-0.16958,0.19978,218.27,0.10501,-0.44821,-0.88774,204.29,0.24009,0.8777,-0.41474,262.15,#3.1,1,0,0,0,0,1,0,0,0,0,1,0,#3.2,1,0,0,0,0,1,0,0,0,0,1,0,#6,-0.014521,-0.06976,0.99746,199.01,0.37884,-0.92358,-0.059077,257.64,0.92535,0.37702,0.039838,302.31,#6.1,1,0,0,0,0,1,0,0,0,0,1,0,#6.2,1,0,0,0,0,1,0,0,0,0,1,0,#6.3,1,0,0,0,0,1,0,0,0,0,1,0,#7,0.8169,-0.21413,0.53556,204.47,0.42791,-0.39761,-0.81167,168.64,0.38675,0.89222,-0.23318,214.6,#7.1,1,0,0,0,0,1,0,0,0,0,1,0,#7.2,1,0,0,0,0,1,0,0,0,0,1,0,#8,0.99431,0.074735,-0.075973,218.33,-0.026887,0.86575,0.49976,177.43,0.10312,-0.49487,0.86283,264.22,#8.1,1,0,0,0,0,1,0,0,0,0,1,0,#8.2,1,0,0,0,0,1,0,0,0,0,1,0,#9,0.296,-0.067032,0.95283,-8.4654,0.64478,-0.72195,-0.25109,250.56,0.70473,0.68869,-0.17048,-85.496,#9.1,1,0,0,0,0,1,0,0,0,0,1,0,#9.2,1,0,0,0,0,1,0,0,0,0,1,0,#9.3,1,0,0,0,0,1,0,0,0,0,1,0,#1,-0.9757,0.21626,0.035289,201.77,-0.14806,-0.76939,0.62138,181.78,0.16153,0.60106,0.78271,185.59,#1.1,1,0,0,0,0,1,0,0,0,0,1,0,#1.2,1,0,0,0,0,1,0,0,0,0,1,0,#1.3,1,0,0,0,0,1,0,0,0,0,1,0
> turn y 90
[Repeated 7 time(s)]
> hide #!9 models
> hide #!8 models
> hide #!7 models
> hide #!6 models
> hide #!3 models
> hide #!2 models
> show #!2 models
> show #!3 models
> show #!6 models
> show #!7 models
> show #!8 models
> show #!9 models
> select add #9
3371 atoms, 3451 bonds, 444 residues, 1 model selected
> select add #8
6711 atoms, 6868 bonds, 883 residues, 4 models selected
> select add #7
10114 atoms, 10352 bonds, 1331 residues, 7 models selected
> select add #6
18996 atoms, 19446 bonds, 1 pseudobond, 2485 residues, 11 models selected
> select add #3
21545 atoms, 22052 bonds, 1 pseudobond, 2816 residues, 14 models selected
> surface style #3,6-9 mesh
> transparency (#!3,6-9 & sel) 30
> transparency (#!3,6-9 & sel) 0
> select clear
> view matrix
view matrix camera
0.056974,0.036338,0.99772,652.72,-0.90561,-0.41878,0.066966,227.85,0.42026,-0.90737,0.009043,200.21
view matrix models
#2,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#2.1,1,0,0,0,0,1,0,0,0,0,1,0,#3,0.96505,-0.16958,0.19978,218.27,0.10501,-0.44821,-0.88774,204.29,0.24009,0.8777,-0.41474,262.15,#3.1,1,0,0,0,0,1,0,0,0,0,1,0,#3.2,1,0,0,0,0,1,0,0,0,0,1,0,#6,-0.014521,-0.06976,0.99746,199.01,0.37884,-0.92358,-0.059077,257.64,0.92535,0.37702,0.039838,302.31,#6.1,1,0,0,0,0,1,0,0,0,0,1,0,#6.2,1,0,0,0,0,1,0,0,0,0,1,0,#6.3,1,0,0,0,0,1,0,0,0,0,1,0,#7,0.8169,-0.21413,0.53556,204.47,0.42791,-0.39761,-0.81167,168.64,0.38675,0.89222,-0.23318,214.6,#7.1,1,0,0,0,0,1,0,0,0,0,1,0,#7.2,1,0,0,0,0,1,0,0,0,0,1,0,#8,0.99431,0.074735,-0.075973,218.33,-0.026887,0.86575,0.49976,177.43,0.10312,-0.49487,0.86283,264.22,#8.1,1,0,0,0,0,1,0,0,0,0,1,0,#8.2,1,0,0,0,0,1,0,0,0,0,1,0,#9,0.296,-0.067032,0.95283,-8.4654,0.64478,-0.72195,-0.25109,250.56,0.70473,0.68869,-0.17048,-85.496,#9.1,1,0,0,0,0,1,0,0,0,0,1,0,#9.2,1,0,0,0,0,1,0,0,0,0,1,0,#9.3,1,0,0,0,0,1,0,0,0,0,1,0,#1,-0.9757,0.21626,0.035289,201.77,-0.14806,-0.76939,0.62138,181.78,0.16153,0.60106,0.78271,185.59,#1.1,1,0,0,0,0,1,0,0,0,0,1,0,#1.2,1,0,0,0,0,1,0,0,0,0,1,0,#1.3,1,0,0,0,0,1,0,0,0,0,1,0
> hide #!2 models
> hide #!3 models
> show #!3 models
> show #!2 models
> hide #!8 models
> hide #!7 models
> hide #!6 models
> hide #!3 models
> show #!3 models
> hide #!3 models
> hide #!2 models
> save 9-4-2024-MODES-Site2-1.png transparentBackground true
> turn y 90
[Repeated 3 time(s)]
> turn x 90
> save 9-4-2024-MODES-Site2-1top.png transparentBackground true
> hide #!9 models
> show #!8 models
> hide #!8 models
> show #!7 models
> turn x 90
[Repeated 2 time(s)]
> turn y 90
[Repeated 2 time(s)]
> show #!3 models
> hide #!3 models
> open "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Structure Files/Gc+Site2+Site5/Pymol/Gc-42437-36124_refine_035.pdb"
Chain information for Gc-42437-36124_refine_035.pdb #4
---
Chain | Description
A | No description available
B | No description available
G | No description available
H | No description available
L | No description available
> mmaker #4 t o #6
> matchmaker #4 to o #6
Invalid "to" argument: invalid atoms specifier
> show #!6 models
> hide #!7 models
> hide #!4 models
> show #!4 models
> mmaker #6 to #4
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker Gc-42437-36124_refine_035.pdb, chain G (#4) with 36124 Fab crystal,
chain G (#6), sequence alignment score = 2561.7
RMSD between 481 pruned atom pairs is 0.000 angstroms; (across all 481 pairs:
0.000)
> hide #!4 models
> hide #!6 models
> hide #!7 models
> show #!2 models
> hide #!1 models
> show #!1 models
> hide #!2 models
> hide #!1 models
> show #!1 models
> hide #!1 models
> show #!2 models
> hide #!2 models
> show #!3 models
> hide #!3 models
> show #!4 models
> mmaker #4 to #6
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker 36124 Fab crystal, chain G (#6) with Gc-42437-36124_refine_035.pdb,
chain G (#4), sequence alignment score = 2561.7
RMSD between 481 pruned atom pairs is 0.000 angstroms; (across all 481 pairs:
0.000)
> hide #!2 models
> show #!4 models
> show #!6 models
> hide #!6 models
> hide #!4 models
> mmaker #4 to #2 bring #6
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker Gc DI-DII from Gc/2/5xstal, chain G (#2) with
Gc-42437-36124_refine_035.pdb, chain G (#4), sequence alignment score = 1511.8
RMSD between 235 pruned atom pairs is 0.897 angstroms; (across all 321 pairs:
2.280)
> show #!2 models
> show #!4 models
> show #!6 models
> hide #!6 models
> hide #!2 models
Drag select of 521 atoms
> select up
705 atoms, 712 bonds, 94 residues, 1 model selected
> select up
1121 atoms, 1137 bonds, 152 residues, 1 model selected
> select up
3361 atoms, 3442 bonds, 443 residues, 1 model selected
> hide sel target a
Drag select of 440 atoms
> select up
606 atoms, 602 bonds, 78 residues, 1 model selected
> select up
1080 atoms, 1101 bonds, 139 residues, 1 model selected
> select up
1714 atoms, 1754 bonds, 226 residues, 1 model selected
> select up
8882 atoms, 9094 bonds, 1154 residues, 1 model selected
> select down
1714 atoms, 1754 bonds, 226 residues, 1 model selected
> hide sel target a
Drag select of 3807 atoms, 1 pseudobonds, 53 bonds
> hide (#!4 & sel) target a
> cartoon (#!4 & sel)
> select clear
> select #4/G: 1041-1140, 1232-1268, 1419-1430
963 atoms, 974 bonds, 127 residues, 1 model selected
> color sel #FF7F7F
> select #4/G: 1141-1186, 1205-1231, 1269-1282, 1296-1320, 1347-1403,
> 1410-1418, 1187-1204
1570 atoms, 1615 bonds, 196 residues, 1 model selected
> color sel #FFFF7F
> select #4/G: 1283-1295, 1321-1346, 1404-1409
331 atoms, 339 bonds, 1 pseudobond, 41 residues, 2 models selected
> color sel #c18c7f
> select #4/G: 1431-1441
90 atoms, 90 bonds, 11 residues, 1 model selected
> color sel #7FFFFF
> select #4/G: 1442-1546
795 atoms, 807 bonds, 105 residues, 1 model selected
> color sel #7F7FFF
> select #4/G: 1547-1572
46 atoms, 48 bonds, 4 residues, 1 model selected
> color sel #FF7FFF
> select add #4
8882 atoms, 9094 bonds, 1 pseudobond, 1154 residues, 2 models selected
> select subtract #4
Nothing selected
> show #!2 models
> show #!3 models
> hide #!3 models
> hide #!2 models
> show #!1 models
> hide #!1 models
> show #!1 models
> hide #!1 models
> show #!8 models
> hide #!8 models
> show #!8 models
> hide #!8 models
> show #!8 models
> show #!6 models
> hide #!8 models
> show #!1 models
> view matrix camera
> 0.056974,0.036338,0.99772,652.72,-0.90561,-0.41878,0.066966,227.85,0.42026,-0.90737,0.009043,200.21
> view matrix models
> #2,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#2.1,1,0,0,0,0,1,0,0,0,0,1,0,#3,0.96505,-0.16958,0.19978,218.27,0.10501,-0.44821,-0.88774,204.29,0.24009,0.8777,-0.41474,262.15,#3.1,1,0,0,0,0,1,0,0,0,0,1,0,#3.2,1,0,0,0,0,1,0,0,0,0,1,0,#6,-0.014521,-0.06976,0.99746,199.01,0.37884,-0.92358,-0.059077,257.64,0.92535,0.37702,0.039838,302.31,#6.1,1,0,0,0,0,1,0,0,0,0,1,0,#6.2,1,0,0,0,0,1,0,0,0,0,1,0,#6.3,1,0,0,0,0,1,0,0,0,0,1,0,#7,0.8169,-0.21413,0.53556,204.47,0.42791,-0.39761,-0.81167,168.64,0.38675,0.89222,-0.23318,214.6,#7.1,1,0,0,0,0,1,0,0,0,0,1,0,#7.2,1,0,0,0,0,1,0,0,0,0,1,0,#8,0.99431,0.074735,-0.075973,218.33,-0.026887,0.86575,0.49976,177.43,0.10312,-0.49487,0.86283,264.22,#8.1,1,0,0,0,0,1,0,0,0,0,1,0,#8.2,1,0,0,0,0,1,0,0,0,0,1,0,#9,0.296,-0.067032,0.95283,-8.4654,0.64478,-0.72195,-0.25109,250.56,0.70473,0.68869,-0.17048,-85.496,#9.1,1,0,0,0,0,1,0,0,0,0,1,0,#9.2,1,0,0,0,0,1,0,0,0,0,1,0,#9.3,1,0,0,0,0,1,0,0,0,0,1,0,#1,-0.9757,0.21626,0.035289,201.77,-0.14806,-0.76939,0.62138,181.78,0.16153,0.60106,0.78271,185.59,#1.1,1,0,0,0,0,1,0,0,0,0,1,0,#1.2,1,0,0,0,0,1,0,0,0,0,1,0,#1.3,1,0,0,0,0,1,0,0,0,0,1,0
> save 9-4-2024-MODES-Site5-1.png transparentBackground true
> turn y 90
[Repeated 2 time(s)]
> select add #6
8882 atoms, 9094 bonds, 1 pseudobond, 1154 residues, 2 models selected
> surface style #6 solid
> select subtract #6
2 models selected
> turn y 90
> view matrix camera
> 0.056974,0.036338,0.99772,652.72,-0.90561,-0.41878,0.066966,227.85,0.42026,-0.90737,0.009043,200.21
> view matrix models
> #2,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#2.1,1,0,0,0,0,1,0,0,0,0,1,0,#3,0.96505,-0.16958,0.19978,218.27,0.10501,-0.44821,-0.88774,204.29,0.24009,0.8777,-0.41474,262.15,#3.1,1,0,0,0,0,1,0,0,0,0,1,0,#3.2,1,0,0,0,0,1,0,0,0,0,1,0,#6,-0.014521,-0.06976,0.99746,199.01,0.37884,-0.92358,-0.059077,257.64,0.92535,0.37702,0.039838,302.31,#6.1,1,0,0,0,0,1,0,0,0,0,1,0,#6.2,1,0,0,0,0,1,0,0,0,0,1,0,#6.3,1,0,0,0,0,1,0,0,0,0,1,0,#7,0.8169,-0.21413,0.53556,204.47,0.42791,-0.39761,-0.81167,168.64,0.38675,0.89222,-0.23318,214.6,#7.1,1,0,0,0,0,1,0,0,0,0,1,0,#7.2,1,0,0,0,0,1,0,0,0,0,1,0,#8,0.99431,0.074735,-0.075973,218.33,-0.026887,0.86575,0.49976,177.43,0.10312,-0.49487,0.86283,264.22,#8.1,1,0,0,0,0,1,0,0,0,0,1,0,#8.2,1,0,0,0,0,1,0,0,0,0,1,0,#9,0.296,-0.067032,0.95283,-8.4654,0.64478,-0.72195,-0.25109,250.56,0.70473,0.68869,-0.17048,-85.496,#9.1,1,0,0,0,0,1,0,0,0,0,1,0,#9.2,1,0,0,0,0,1,0,0,0,0,1,0,#9.3,1,0,0,0,0,1,0,0,0,0,1,0,#1,-0.9757,0.21626,0.035289,201.77,-0.14806,-0.76939,0.62138,181.78,0.16153,0.60106,0.78271,185.59,#1.1,1,0,0,0,0,1,0,0,0,0,1,0,#1.2,1,0,0,0,0,1,0,0,0,0,1,0,#1.3,1,0,0,0,0,1,0,0,0,0,1,0
> show #!7 models
> show #!8 models
> show #!9 models
> show #!3 models
> surface style solid
> select clear
> hide #!8 models
> hide #!7 models
> hide #!6 models
> hide #!4 models
> hide #!3 models
> save 9-4-2024-MODES-Site2-front2.png transparentBackground true
> turn x 90
> save 9-4-2024-MODES-Site2-top2.png transparentBackground true
> turn x 90
[Repeated 2 time(s)]
> hide #!9 models
> show #!8 models
> hide #!8 models
> show #!6 models
> hide #!6 models
> show #!7 models
> turn y 90
[Repeated 1 time(s)]
> show #!3 models
> hide #!3 models
> show #!4 models
> hide #!4 models
> view matrix camera
> 0.056974,0.036338,0.99772,652.72,-0.90561,-0.41878,0.066966,227.85,0.42026,-0.90737,0.009043,200.21
> view matrix models
> #2,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#2.1,1,0,0,0,0,1,0,0,0,0,1,0,#3,0.96505,-0.16958,0.19978,218.27,0.10501,-0.44821,-0.88774,204.29,0.24009,0.8777,-0.41474,262.15,#3.1,1,0,0,0,0,1,0,0,0,0,1,0,#3.2,1,0,0,0,0,1,0,0,0,0,1,0,#6,-0.014521,-0.06976,0.99746,199.01,0.37884,-0.92358,-0.059077,257.64,0.92535,0.37702,0.039838,302.31,#6.1,1,0,0,0,0,1,0,0,0,0,1,0,#6.2,1,0,0,0,0,1,0,0,0,0,1,0,#6.3,1,0,0,0,0,1,0,0,0,0,1,0,#7,0.8169,-0.21413,0.53556,204.47,0.42791,-0.39761,-0.81167,168.64,0.38675,0.89222,-0.23318,214.6,#7.1,1,0,0,0,0,1,0,0,0,0,1,0,#7.2,1,0,0,0,0,1,0,0,0,0,1,0,#8,0.99431,0.074735,-0.075973,218.33,-0.026887,0.86575,0.49976,177.43,0.10312,-0.49487,0.86283,264.22,#8.1,1,0,0,0,0,1,0,0,0,0,1,0,#8.2,1,0,0,0,0,1,0,0,0,0,1,0,#9,0.296,-0.067032,0.95283,-8.4654,0.64478,-0.72195,-0.25109,250.56,0.70473,0.68869,-0.17048,-85.496,#9.1,1,0,0,0,0,1,0,0,0,0,1,0,#9.2,1,0,0,0,0,1,0,0,0,0,1,0,#9.3,1,0,0,0,0,1,0,0,0,0,1,0,#1,-0.9757,0.21626,0.035289,201.77,-0.14806,-0.76939,0.62138,181.78,0.16153,0.60106,0.78271,185.59,#1.1,1,0,0,0,0,1,0,0,0,0,1,0,#1.2,1,0,0,0,0,1,0,0,0,0,1,0,#1.3,1,0,0,0,0,1,0,0,0,0,1,0
> turn y 90
[Repeated 1 time(s)]
> save 9-4-2024-MODES-Site4-front1.png transparentBackground true
> show #!4 models
> hide #!1 models
> save 9-4-2024-MODES-Site4-front2.png transparentBackground true
> hide #!7 models
> show #!6 models
> save 9-4-2024-MODES-Site5-pre1.png transparentBackground true
> show #!1 models
> save 9-4-2024-MODES-Site5-post1.png transparentBackground true
> hide #!6 models
> show #!8 models
> hide #!1 models
> show #!3 models
> hide #!8 models
> show #!2 models
> hide #!4 models
> show #!8 models
> save 9-4-2024-MODES-Site6-pre1.png transparentBackground true
> show #!1 models
> hide #!2 models
> show #!2 models
> hide #!2 models
> turn y 90
[Repeated 1 time(s)]
> show #!2 models
> hide #!2 models
> save 9-4-2024-MODES-Site6-both1.png transparentBackground true
> hide #!1 models
> show #!2 models
> hide #!3 models
> hide #!2 models
> show #!4 models
> show #!6 models
> show #!7 models
> show #!9 models
> view matrix camera
> 0.056974,0.036338,0.99772,652.72,-0.90561,-0.41878,0.066966,227.85,0.42026,-0.90737,0.009043,200.21
> view matrix models
> #2,0.99404,0.10845,-0.011097,-16.565,-0.1089,0.99252,-0.055252,33.615,0.0050223,0.056131,0.99841,-20.521,#2.1,1,0,0,0,0,1,0,0,0,0,1,0,#3,0.96505,-0.16958,0.19978,218.27,0.10501,-0.44821,-0.88774,204.29,0.24009,0.8777,-0.41474,262.15,#3.1,1,0,0,0,0,1,0,0,0,0,1,0,#3.2,1,0,0,0,0,1,0,0,0,0,1,0,#6,-0.014521,-0.06976,0.99746,199.01,0.37884,-0.92358,-0.059077,257.64,0.92535,0.37702,0.039838,302.31,#6.1,1,0,0,0,0,1,0,0,0,0,1,0,#6.2,1,0,0,0,0,1,0,0,0,0,1,0,#6.3,1,0,0,0,0,1,0,0,0,0,1,0,#7,0.8169,-0.21413,0.53556,204.47,0.42791,-0.39761,-0.81167,168.64,0.38675,0.89222,-0.23318,214.6,#7.1,1,0,0,0,0,1,0,0,0,0,1,0,#7.2,1,0,0,0,0,1,0,0,0,0,1,0,#8,0.99431,0.074735,-0.075973,218.33,-0.026887,0.86575,0.49976,177.43,0.10312,-0.49487,0.86283,264.22,#8.1,1,0,0,0,0,1,0,0,0,0,1,0,#8.2,1,0,0,0,0,1,0,0,0,0,1,0,#9,0.296,-0.067032,0.95283,-8.4654,0.64478,-0.72195,-0.25109,250.56,0.70473,0.68869,-0.17048,-85.496,#9.1,1,0,0,0,0,1,0,0,0,0,1,0,#9.2,1,0,0,0,0,1,0,0,0,0,1,0,#9.3,1,0,0,0,0,1,0,0,0,0,1,0,#1,-0.9757,0.21626,0.035289,201.77,-0.14806,-0.76939,0.62138,181.78,0.16153,0.60106,0.78271,185.59,#1.1,1,0,0,0,0,1,0,0,0,0,1,0,#1.2,1,0,0,0,0,1,0,0,0,0,1,0,#1.3,1,0,0,0,0,1,0,0,0,0,1,0'
Invalid "models" argument: Require numeric values
> save "C:/Users/ckh755/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Figures/Supplemental Figure #_modes of neut/9-4-2024_Gc with all 4
> Sites models.cxs"
——— End of log from Wed Sep 4 19:17:24 2024 ———
opened ChimeraX session
> turn y 90
[Repeated 1 time(s)]
> show #!1 models
> hide #!6 models
> hide #!7 models
> hide #!9 models
> show #!3 models
> mmaker #3 to #4 bring #8
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker Gc-42437-36124_refine_035.pdb, chain G (#4) with AF3_domain III
from bound w36145Fv, chain A (#3), sequence alignment score = 520.8
RMSD between 105 pruned atom pairs is 0.570 angstroms; (across all 105 pairs:
0.570)
> open "C:/Users/chris/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Structure Files/Gc-4Ffabs/Map for Gc245 med res map with Site 6 and
> domain III/fold_gc_diii_36145_fv_model_4_Coot-updatednumbering.pdb"
Chain information for fold_gc_diii_36145_fv_model_4_Coot-updatednumbering.pdb
#5
---
Chain | Description
A | No description available
B | No description available
C | No description available
> mmaker #5 to #8
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker AF2 36145 Fab, chain A (#8) with
fold_gc_diii_36145_fv_model_4_Coot-updatednumbering.pdb, chain B (#5),
sequence alignment score = 614.5
RMSD between 118 pruned atom pairs is 0.257 angstroms; (across all 118 pairs:
0.257)
> mmaker #5 to #1/A
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker 7a59, chain A (#1) with fold_gc_diii_36145_fv_model_4_Coot-
updatednumbering.pdb, chain A (#5), sequence alignment score = 500.4
RMSD between 100 pruned atom pairs is 0.595 angstroms; (across all 105 pairs:
1.273)
> hide #!4 models
> hide #!3 models
> hide #!1 models
> hide #!8 models
Drag select of 130 residues
> select up
1357 atoms, 1384 bonds, 175 residues, 1 model selected
> select up
1753 atoms, 1794 bonds, 226 residues, 1 model selected
> color sel #DD833E
> cartoon hide sel
> surface sel
> select clear
Drag select of 41 residues
> select up
475 atoms, 482 bonds, 63 residues, 1 model selected
> select up
796 atoms, 808 bonds, 105 residues, 1 model selected
> color sel #7F7FFF
> select clear
> show #!4 models
> hide #!4 models
> show #!1 models
> show #!2 models
> hide #!2 models
> show #!8 models
> show #!3 models
> hide #!5 models
> save C:\Users\chris/Desktop\image1.png supersample 3
> show #!5 models
> save C:\Users\chris/Desktop\image2.png supersample 3
> turn y 90
> save C:\Users\chris/Desktop\image3.png supersample 3
> save C:\Users\chris/Desktop\image4.png supersample 3
> show #!7 models
> hide #!7 models
> hide #!3 models
> show #!3 models
> save "C:/Users/chris/Box/Christy Hjorth/CCHFV/Manuscripts/Gc Ab
> Paper/Structure Files/Gc-4Ffabs/Map for Gc245 med res map with Site 6 and
> domain III/9-23-2024_36145 and domain III.cxs"
Traceback (most recent call last):
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\core\triggerset.py", line 149, in invoke
return self._func(self._name, data)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\ui\file_history.py", line 93, in file_history_changed_cb
self.update_html()
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\ui\file_history.py", line 88, in update_html
fhw.setHtml(html)
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\ui\widgets\htmlview.py", line 244, in setHtml
tf = open(self._tf_name, "wb")
^^^^^^^^^^^^^^^^^^^^^^^^^
PermissionError: [Errno 13] Permission denied:
'C:\\\Users\\\chris\\\AppData\\\Local\\\Temp\\\chbp8qmgij_1.html'
Error processing trigger "file history changed":
PermissionError: [Errno 13] Permission denied:
'C:\\\Users\\\chris\\\AppData\\\Local\\\Temp\\\chbp8qmgij_1.html'
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\ui\widgets\htmlview.py", line 244, in setHtml
tf = open(self._tf_name, "wb")
^^^^^^^^^^^^^^^^^^^^^^^^^
See log for complete Python traceback.
Traceback (most recent call last):
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\core\triggerset.py", line 149, in invoke
return self._func(self._name, data)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\ui\file_history.py", line 93, in file_history_changed_cb
self.update_html()
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\ui\file_history.py", line 88, in update_html
fhw.setHtml(html)
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\ui\widgets\htmlview.py", line 244, in setHtml
tf = open(self._tf_name, "wb")
^^^^^^^^^^^^^^^^^^^^^^^^^
PermissionError: [Errno 13] Permission denied:
'C:\\\Users\\\chris\\\AppData\\\Local\\\Temp\\\chbp8qmgij_1.html'
Error processing trigger "file history changed":
PermissionError: [Errno 13] Permission denied:
'C:\\\Users\\\chris\\\AppData\\\Local\\\Temp\\\chbp8qmgij_1.html'
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\ui\widgets\htmlview.py", line 244, in setHtml
tf = open(self._tf_name, "wb")
^^^^^^^^^^^^^^^^^^^^^^^^^
See log for complete Python traceback.
> info path user unversioned data
user unversioned data directory: C:\Users\chris\AppData\Local\UCSF\ChimeraX
> cd C:/Apps/ChimeraX
Current working directory is: C:\Apps\ChimeraX
> save C:/Apps/ChimeraX/test.cxs
Traceback (most recent call last):
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\core\triggerset.py", line 149, in invoke
return self._func(self._name, data)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\ui\file_history.py", line 93, in file_history_changed_cb
self.update_html()
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\ui\file_history.py", line 88, in update_html
fhw.setHtml(html)
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\ui\widgets\htmlview.py", line 244, in setHtml
tf = open(self._tf_name, "wb")
^^^^^^^^^^^^^^^^^^^^^^^^^
PermissionError: [Errno 13] Permission denied:
'C:\\\Users\\\chris\\\AppData\\\Local\\\Temp\\\chbp8qmgij_1.html'
Error processing trigger "file history changed":
PermissionError: [Errno 13] Permission denied:
'C:\\\Users\\\chris\\\AppData\\\Local\\\Temp\\\chbp8qmgij_1.html'
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\ui\widgets\htmlview.py", line 244, in setHtml
tf = open(self._tf_name, "wb")
^^^^^^^^^^^^^^^^^^^^^^^^^
See log for complete Python traceback.
Traceback (most recent call last):
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\core\triggerset.py", line 149, in invoke
return self._func(self._name, data)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\ui\file_history.py", line 93, in file_history_changed_cb
self.update_html()
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\ui\file_history.py", line 88, in update_html
fhw.setHtml(html)
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\ui\widgets\htmlview.py", line 244, in setHtml
tf = open(self._tf_name, "wb")
^^^^^^^^^^^^^^^^^^^^^^^^^
PermissionError: [Errno 13] Permission denied:
'C:\\\Users\\\chris\\\AppData\\\Local\\\Temp\\\chbp8qmgij_1.html'
Error processing trigger "file history changed":
PermissionError: [Errno 13] Permission denied:
'C:\\\Users\\\chris\\\AppData\\\Local\\\Temp\\\chbp8qmgij_1.html'
File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-
packages\chimerax\ui\widgets\htmlview.py", line 244, in setHtml
tf = open(self._tf_name, "wb")
^^^^^^^^^^^^^^^^^^^^^^^^^
See log for complete Python traceback.
OpenGL version: 3.3.0 NVIDIA 512.72
OpenGL renderer: NVIDIA GeForce GTX 1650 Ti/PCIe/SSE2
OpenGL vendor: NVIDIA Corporation
Python: 3.11.4
Locale: en_US.cp1252
Qt version: PyQt6 6.6.1, Qt 6.6.1
Qt runtime version: 6.6.3
Qt platform: windows
Manufacturer: Dell Inc.
Model: G3 3500
OS: Microsoft Windows 11 Home (Build 22631)
Memory: 16,963,534,848
MaxProcessMemory: 137,438,953,344
CPU: 12 Intel(R) Core(TM) i7-10750H CPU @ 2.60GHz
OSLanguage: en-US
Installed Packages:
alabaster: 0.7.16
appdirs: 1.4.4
asttokens: 2.4.1
Babel: 2.15.0
beautifulsoup4: 4.12.3
blockdiag: 3.0.0
blosc2: 2.0.0
build: 1.2.1
certifi: 2024.6.2
cftime: 1.6.4
charset-normalizer: 3.3.2
ChimeraX-AddCharge: 1.5.17
ChimeraX-AddH: 2.2.6
ChimeraX-AlignmentAlgorithms: 2.0.2
ChimeraX-AlignmentHdrs: 3.5
ChimeraX-AlignmentMatrices: 2.1
ChimeraX-Alignments: 2.12.7
ChimeraX-AlphaFold: 1.0
ChimeraX-AltlocExplorer: 1.1.1
ChimeraX-AmberInfo: 1.0
ChimeraX-Arrays: 1.1
ChimeraX-Atomic: 1.57.1
ChimeraX-AtomicLibrary: 14.0.6
ChimeraX-AtomSearch: 2.0.1
ChimeraX-AxesPlanes: 2.4
ChimeraX-BasicActions: 1.1.2
ChimeraX-BILD: 1.0
ChimeraX-BlastProtein: 2.4.6
ChimeraX-BondRot: 2.0.4
ChimeraX-BugReporter: 1.0.1
ChimeraX-BuildStructure: 2.12.1
ChimeraX-Bumps: 1.0
ChimeraX-BundleBuilder: 1.2.7
ChimeraX-ButtonPanel: 1.0.1
ChimeraX-CageBuilder: 1.0.1
ChimeraX-CellPack: 1.0
ChimeraX-Centroids: 1.4
ChimeraX-ChangeChains: 1.1
ChimeraX-CheckWaters: 1.4
ChimeraX-ChemGroup: 2.0.1
ChimeraX-Clashes: 2.2.4
ChimeraX-Clipper: 0.23.1
ChimeraX-ColorActions: 1.0.5
ChimeraX-ColorGlobe: 1.0
ChimeraX-ColorKey: 1.5.6
ChimeraX-CommandLine: 1.2.5
ChimeraX-ConnectStructure: 2.0.1
ChimeraX-Contacts: 1.0.1
ChimeraX-Core: 1.8
ChimeraX-CoreFormats: 1.2
ChimeraX-coulombic: 1.4.3
ChimeraX-Crosslinks: 1.0
ChimeraX-Crystal: 1.0
ChimeraX-CrystalContacts: 1.0.1
ChimeraX-DataFormats: 1.2.3
ChimeraX-Dicom: 1.2.4
ChimeraX-DiffPlot: 1.0
ChimeraX-DistMonitor: 1.4.2
ChimeraX-DockPrep: 1.1.3
ChimeraX-Dssp: 2.0
ChimeraX-EMDB-SFF: 1.0
ChimeraX-ESMFold: 1.0
ChimeraX-FileHistory: 1.0.1
ChimeraX-FunctionKey: 1.0.1
ChimeraX-Geometry: 1.3
ChimeraX-gltf: 1.0
ChimeraX-Graphics: 1.1.1
ChimeraX-Hbonds: 2.4
ChimeraX-Help: 1.2.2
ChimeraX-HKCage: 1.3
ChimeraX-IHM: 1.1
ChimeraX-ImageFormats: 1.2
ChimeraX-IMOD: 1.0
ChimeraX-IO: 1.0.1
ChimeraX-ISOLDE: 1.8
ChimeraX-ItemsInspection: 1.0.1
ChimeraX-IUPAC: 1.0
ChimeraX-Label: 1.1.10
ChimeraX-ListInfo: 1.2.2
ChimeraX-Log: 1.1.6
ChimeraX-LookingGlass: 1.1
ChimeraX-Maestro: 1.9.1
ChimeraX-Map: 1.2
ChimeraX-MapData: 2.0
ChimeraX-MapEraser: 1.0.1
ChimeraX-MapFilter: 2.0.1
ChimeraX-MapFit: 2.0
ChimeraX-MapSeries: 2.1.1
ChimeraX-Markers: 1.0.1
ChimeraX-Mask: 1.0.2
ChimeraX-MatchMaker: 2.1.3
ChimeraX-MCopy: 1.0
ChimeraX-MDcrds: 2.7.1
ChimeraX-MedicalToolbar: 1.0.3
ChimeraX-Meeting: 1.0.1
ChimeraX-MLP: 1.1.1
ChimeraX-mmCIF: 2.14.1
ChimeraX-MMTF: 2.2
ChimeraX-Modeller: 1.5.17
ChimeraX-ModelPanel: 1.5
ChimeraX-ModelSeries: 1.0.1
ChimeraX-Mol2: 2.0.3
ChimeraX-Mole: 1.0
ChimeraX-Morph: 1.0.2
ChimeraX-MouseModes: 1.2
ChimeraX-Movie: 1.0
ChimeraX-Neuron: 1.0
ChimeraX-Nifti: 1.2
ChimeraX-NMRSTAR: 1.0.2
ChimeraX-NRRD: 1.2
ChimeraX-Nucleotides: 2.0.3
ChimeraX-OpenCommand: 1.13.5
ChimeraX-PDB: 2.7.5
ChimeraX-PDBBio: 1.0.1
ChimeraX-PDBLibrary: 1.0.4
ChimeraX-PDBMatrices: 1.0
ChimeraX-PickBlobs: 1.0.1
ChimeraX-Positions: 1.0
ChimeraX-PresetMgr: 1.1.1
ChimeraX-PubChem: 2.2
ChimeraX-ReadPbonds: 1.0.1
ChimeraX-Registration: 1.1.2
ChimeraX-RemoteControl: 1.0
ChimeraX-RenderByAttr: 1.4.1
ChimeraX-RenumberResidues: 1.1
ChimeraX-ResidueFit: 1.0.1
ChimeraX-RestServer: 1.2
ChimeraX-RNALayout: 1.0
ChimeraX-RotamerLibMgr: 4.0
ChimeraX-RotamerLibsDunbrack: 2.0
ChimeraX-RotamerLibsDynameomics: 2.0
ChimeraX-RotamerLibsRichardson: 2.0
ChimeraX-SaveCommand: 1.5.1
ChimeraX-SchemeMgr: 1.0
ChimeraX-SDF: 2.0.2
ChimeraX-Segger: 1.0
ChimeraX-Segment: 1.0.1
ChimeraX-Segmentations: 3.0.15
ChimeraX-SelInspector: 1.0
ChimeraX-SeqView: 2.11.2
ChimeraX-Shape: 1.0.1
ChimeraX-Shell: 1.0.1
ChimeraX-Shortcuts: 1.1.1
ChimeraX-ShowSequences: 1.0.3
ChimeraX-SideView: 1.0.1
ChimeraX-Smiles: 2.1.2
ChimeraX-SmoothLines: 1.0
ChimeraX-SpaceNavigator: 1.0
ChimeraX-StdCommands: 1.16.5
ChimeraX-STL: 1.0.1
ChimeraX-Storm: 1.0
ChimeraX-StructMeasure: 1.2.1
ChimeraX-Struts: 1.0.1
ChimeraX-Surface: 1.0.1
ChimeraX-SwapAA: 2.0.1
ChimeraX-SwapRes: 2.5
ChimeraX-TapeMeasure: 1.0
ChimeraX-TaskManager: 1.0
ChimeraX-Test: 1.0
ChimeraX-Toolbar: 1.1.2
ChimeraX-ToolshedUtils: 1.2.4
ChimeraX-Topography: 1.0
ChimeraX-ToQuest: 1.0
ChimeraX-Tug: 1.0.1
ChimeraX-UI: 1.39.1
ChimeraX-uniprot: 2.3
ChimeraX-UnitCell: 1.0.1
ChimeraX-ViewDockX: 1.4.3
ChimeraX-VIPERdb: 1.0
ChimeraX-Vive: 1.1
ChimeraX-VolumeMenu: 1.0.1
ChimeraX-vrml: 1.0
ChimeraX-VTK: 1.0
ChimeraX-WavefrontOBJ: 1.0
ChimeraX-WebCam: 1.0.2
ChimeraX-WebServices: 1.1.4
ChimeraX-Zone: 1.0.1
colorama: 0.4.6
comm: 0.2.2
comtypes: 1.4.1
contourpy: 1.2.1
cxservices: 1.2.2
cycler: 0.12.1
Cython: 3.0.10
debugpy: 1.8.1
decorator: 5.1.1
docutils: 0.20.1
executing: 2.0.1
filelock: 3.13.4
fonttools: 4.53.0
funcparserlib: 2.0.0a0
glfw: 2.7.0
grako: 3.16.5
h5py: 3.11.0
html2text: 2024.2.26
idna: 3.7
ihm: 1.0
imagecodecs: 2024.1.1
imagesize: 1.4.1
ipykernel: 6.29.2
ipython: 8.21.0
ipywidgets: 8.1.3
jedi: 0.19.1
jinja2: 3.1.4
jupyter-client: 8.6.0
jupyter-core: 5.7.2
jupyterlab-widgets: 3.0.11
kiwisolver: 1.4.5
line-profiler: 4.1.2
lxml: 5.2.1
lz4: 4.3.3
MarkupSafe: 2.1.5
matplotlib: 3.8.4
matplotlib-inline: 0.1.7
msgpack: 1.0.8
nest-asyncio: 1.6.0
netCDF4: 1.6.5
networkx: 3.3
nibabel: 5.2.0
nptyping: 2.5.0
numexpr: 2.10.0
numpy: 1.26.4
openvr: 1.26.701
packaging: 24.1
ParmEd: 4.2.2
parso: 0.8.4
pep517: 0.13.1
pillow: 10.3.0
pip: 24.0
pkginfo: 1.10.0
platformdirs: 4.2.2
prompt-toolkit: 3.0.47
psutil: 5.9.8
pure-eval: 0.2.2
py-cpuinfo: 9.0.0
pycollada: 0.8
pydicom: 2.4.4
pygments: 2.17.2
pynmrstar: 3.3.4
pynrrd: 1.0.0
PyOpenGL: 3.1.7
PyOpenGL-accelerate: 3.1.7
pyopenxr: 1.0.3401
pyparsing: 3.1.2
pyproject-hooks: 1.1.0
PyQt6-commercial: 6.6.1
PyQt6-Qt6: 6.6.3
PyQt6-sip: 13.6.0
PyQt6-WebEngine-commercial: 6.6.0
PyQt6-WebEngine-Qt6: 6.6.3
python-dateutil: 2.9.0.post0
pytz: 2024.1
pywin32: 306
pyzmq: 26.0.3
qtconsole: 5.5.1
QtPy: 2.4.1
RandomWords: 0.4.0
requests: 2.31.0
scipy: 1.13.0
setuptools: 69.5.1
sfftk-rw: 0.8.1
six: 1.16.0
snowballstemmer: 2.2.0
sortedcontainers: 2.4.0
soupsieve: 2.5
sphinx: 7.2.6
sphinx-autodoc-typehints: 2.0.1
sphinxcontrib-applehelp: 1.0.8
sphinxcontrib-blockdiag: 3.0.0
sphinxcontrib-devhelp: 1.0.6
sphinxcontrib-htmlhelp: 2.0.5
sphinxcontrib-jsmath: 1.0.1
sphinxcontrib-qthelp: 1.0.7
sphinxcontrib-serializinghtml: 1.1.10
stack-data: 0.6.3
superqt: 0.6.3
tables: 3.8.0
tcia-utils: 1.5.1
tifffile: 2024.1.30
tinyarray: 1.2.4
tornado: 6.4.1
traitlets: 5.14.2
typing-extensions: 4.12.2
tzdata: 2024.1
urllib3: 2.2.1
wcwidth: 0.2.13
webcolors: 1.13
wheel: 0.43.0
wheel-filename: 1.4.1
widgetsnbextension: 4.0.11
WMI: 1.5.1
File attachment: ChimeraX Save Issue.txt
Attachments (3)
Change History (9)
by , 17 months ago
| Attachment: | ChimeraX Save Issue.txt added |
|---|
comment:1 by , 17 months ago
| Cc: | added |
|---|---|
| Component: | Unassigned → UI |
| Owner: | set to |
| Platform: | → all |
| Project: | → ChimeraX |
| Status: | new → accepted |
| Summary: | ChimeraX bug report submission → Cannot update file history |
comment:2 by , 17 months ago
| Status: | accepted → feedback |
|---|
Hi Christy,
Thanks for reporting this problem. So I believe your actual session files are saving correctly, it's the update of the "recently used files" list that is failing. It is trying to write a file into C:\Users\chris\AppData\Local\Temp, which you *should* have permission for. Using the Windows File Explorer, click on its View menu and click the "Hidden" check box. Then navigate to C:\Users\chris\AppData\Local. Right click on the "Temp" entry and choose "Properties". Go to the Property panel's Security tab. Does it say that you have write permission? If it does, click on the "Advanced" button under that. In the Advanced panel does it say that you are the owner? Are you in the permissions lists in that panel?
--Eric
Eric Pettersen
UCSF Computer Graphics Lab
comment:3 by , 17 months ago
Hi Eric, Thanks for prompt reply! I tested a saved session and it indeed opens as expected. Following those steps above, I navigated to the \Local\Temp folder and confirmed that I have writing permission. My user account has the same permissions as the listed admin (also me since personal laptop and not university IT issue) and my account is listed as the Owner. I checked the chbpu99nm_40.html file that was listed as the issue and saw that the file was updated to my most recent save and ChimeraX shows the updated Recently Used Session Files grid. So I'm not quite sure why it has that error popping up if that file is updated and seen within ChimeraX too. I navigated to the other file mentioned: File "C:\Program Files\ChimeraX 1.8\bin\Lib\site-packages\chimerax\ui\widgets\htmlview.py In the chimerax - Properties - Security panel, there are more user names listed: [image: image.png] [image: image.png] Looks like the Users group doesn't have Write permissions here. Should I edit these permissions? Best, *Christy Hjorth* PhD Candidate | McLellan Lab Department of Molecular Biosciences College of Natural Sciences The University of Texas at Austin On Mon, Sep 23, 2024 at 3:09 PM ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu> wrote: > > > > > >
comment:4 by , 17 months ago
Hi Christy,
If you don't have any ChimeraXs running, are there any chbp<stuff>.html files in that folder? If there are, delete them. Then run ChimeraX, and when you get the error see if the file it complains about has shown up in that folder.
The htmlview.py file is just the source code trying to write the temp file. There is no reason you would need to have writer permission on the source code file.
--Eric
comment:5 by , 17 months ago
Hi Eric and Pett, Without ChimeraX running, there are no other chbp...html files in that Local\Temp folder. When I run ChimeraX, I see the chbp...html file appear back in the folder. Best, *Christy Hjorth* PhD Candidate | McLellan Lab Department of Molecular Biosciences College of Natural Sciences The University of Texas at Austin On Mon, Sep 23, 2024 at 7:09 PM ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu> wrote: > > >
comment:6 by , 17 months ago
Hi Christy,
Is your machine running anti-virus software? If so, can you turn it off temporarily and see if that affects the behavior?
--Eric
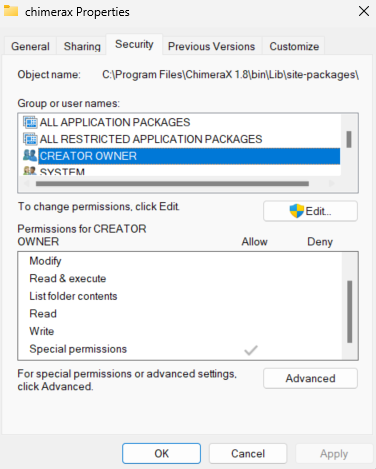
Added by email2trac