Opened 21 months ago
Closed 21 months ago
#15428 closed defect (fixed)
problem with viruses bychain outputs in flows
| Reported by: | Owned by: | Eric Pettersen | |
|---|---|---|---|
| Priority: | normal | Milestone: | |
| Component: | Depiction | Version: | |
| Keywords: | Cc: | Tom Goddard, phil.cruz@…, Bhinnata.piya@… | |
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
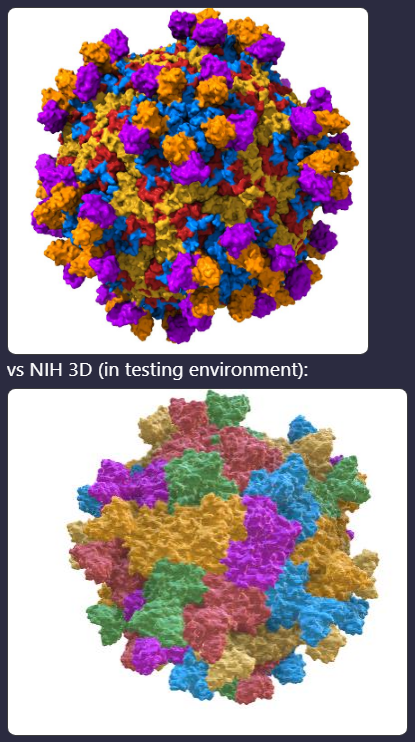
Hey all! I just stumbled into this one today. I was looking at PDB 6WDT and there is an inconsistency between how the preset comes out in ChimeraX vs NIH 3D: ChimeraX: [cid:image001.png@01DABD93.19087DC0] The ChimeraX preset is how I would expect it to look with each chain of the sym units a different color. But when this comes out of NIH 3D using our current prod and our most up-to-date testing environment it looks like the below where each sym unit seems to be colored differently instead of by-chain. I checked with Phil and this isn't what we would expect, so was hoping that Eric could look into it? Thanks! Kristen
Attachments (1)
Change History (7)
by , 21 months ago
| Attachment: | image001.png added |
|---|
comment:1 by , 21 months ago
| Cc: | added |
|---|---|
| Component: | Unassigned → Depiction |
| Owner: | set to |
| Platform: | → all |
| Project: | → ChimeraX |
| Status: | new → accepted |
Hi Kristen,
The issue is that the pipeline script opens the structure with "open pdbe_bio:6wdt maxassemblies 1", which produces chain IDs of A, AA, AAA, through ABG for the 60 copies of chain A from the original asymmetric unit. Similarly for chains B-F of the asymmetric unit. Whereas if you simply open 6wdt and click on the first entry of its assembly table, you get 60 chain As, 60 chain Bs, etc. These 2 sets of chain IDs color differently when you "rainbow chains" on them (which is what the preset is doing).
If you look at the pipeline "color bypolymer" output for 6wdt you will see that the color arrangement is the same as the "by chain" you did by hand (though the colors themselves will be different).
So the question is what do we want to do about this. There seems to be metadata in the pdbe_bio file indicating that it is an assembly, so it's possible that the by-chains preset could do something different/smarter if it detects that.
--Eric
comment:2 by , 21 months ago
This is definitely a Phil question so I’ll let him rely.
Get Outlook for iOS<https://aka.ms/o0ukef>
________________________________
From: ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu>
Sent: Thursday, June 13, 2024 6:30:53 PM
To: Browne, Kristen (NIH/NIAID) [C] <kristen.browne@nih.gov>; pett@cgl.ucsf.edu <pett@cgl.ucsf.edu>
Cc: goddard@cgl.ucsf.edu <goddard@cgl.ucsf.edu>; Cruz, Phil (NIH/NIAID) [C] <phil.cruz@nih.gov>
Subject: [EXTERNAL] Re: [ChimeraX] #15428: problem with viruses bychain outputs in flows
#15428: problem with viruses bychain outputs in flows
---------------------------------------+----------------------
Reporter: kristen.browne@… | Owner: pett
Type: defect | Status: accepted
Priority: normal | Milestone:
Component: Depiction | Version:
Resolution: | Keywords:
Blocked By: | Blocking:
Notify when closed: | Platform: all
Project: ChimeraX |
---------------------------------------+----------------------
Changes (by pett):
* cc: Tom Goddard, phil.cruz@… (added)
* component: Unassigned => Depiction
* owner: (none) => pett
* platform: => all
* project: => ChimeraX
* status: new => accepted
Comment:
Hi Kristen,
The issue is that the pipeline script opens the structure with
"open pdbe_bio:6wdt maxassemblies 1", which produces chain IDs of A, AA,
AAA, through ABG for the 60 copies of chain A from the original asymmetric
unit. Similarly for chains B-F of the asymmetric unit. Whereas if you
simply open 6wdt and click on the first entry of its assembly table, you
get 60 chain As, 60 chain Bs, etc. These 2 sets of chain IDs color
differently when you "rainbow chains" on them (which is what the preset is
doing).
If you look at the pipeline "color bypolymer" output for 6wdt you
will see that the color arrangement is the same as the "by chain" you did
by hand (though the colors themselves will be different).
So the question is what do we want to do about this. There seems
to be metadata in the pdbe_bio file indicating that it is an assembly, so
it's possible that the by-chains preset could do something
different/smarter if it detects that.
--Eric
--
Ticket URL: <https://gcc02.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.rbvi.ucsf.edu%2Ftrac%2FChimeraX%2Fticket%2F15428%23comment%3A1&data=05%7C02%7Ckristen.browne%40nih.gov%7Cfbef6798674d41076f1208dc8bf881f1%7C14b77578977342d58507251ca2dc2b06%7C0%7C0%7C638539146661168344%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C0%7C%7C%7C&sdata=66sQ8jCUQtXL3BHFrjqhk1TmaKFRMNC1rbWixJpQ5Tk%3D&reserved=0<https://www.rbvi.ucsf.edu/trac/ChimeraX/ticket/15428#comment:1>>
ChimeraX <https://gcc02.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.rbvi.ucsf.edu%2Fchimerax%2F&data=05%7C02%7Ckristen.browne%40nih.gov%7Cfbef6798674d41076f1208dc8bf881f1%7C14b77578977342d58507251ca2dc2b06%7C0%7C0%7C638539146661178042%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C0%7C%7C%7C&sdata=2kbrrWiHPtchyLXJYsrQuyRSs8heEUUuQcEgH%2FexUc4%3D&reserved=0<https://www.rbvi.ucsf.edu/chimerax/>>
ChimeraX Issue Tracker
CAUTION: This email originated from outside of the organization. Do not click links or open attachments unless you recognize the sender and are confident the content is safe.
comment:3 by , 21 months ago
Let’s also add bhinnata to this thread so she can track the issue. Bhinnata.piya@nih.gov
Get Outlook for iOS<https://aka.ms/o0ukef>
________________________________
From: ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu>
Sent: Thursday, June 13, 2024 6:54:48 PM
To: pett@cgl.ucsf.edu <pett@cgl.ucsf.edu>; Browne, Kristen (NIH/NIAID) [C] <kristen.browne@nih.gov>
Cc: goddard@cgl.ucsf.edu <goddard@cgl.ucsf.edu>; Cruz, Phil (NIH/NIAID) [C] <phil.cruz@nih.gov>
Subject: [EXTERNAL] Re: [ChimeraX] #15428: problem with viruses bychain outputs in flows
#15428: problem with viruses bychain outputs in flows
---------------------------------------+----------------------
Reporter: kristen.browne@… | Owner: pett
Type: defect | Status: accepted
Priority: normal | Milestone:
Component: Depiction | Version:
Resolution: | Keywords:
Blocked By: | Blocking:
Notify when closed: | Platform: all
Project: ChimeraX |
---------------------------------------+----------------------
Comment (by kristen.browne@…):
{{{
This is definitely a Phil question so I’ll let him rely.
Get Outlook for iOS<https://gcc02.safelinks.protection.outlook.com/?url=https%3A%2F%2Faka.ms%2Fo0ukef&data=05%7C02%7Ckristen.browne%40nih.gov%7C22ff71f196094e8e42c008dc8bfbd683%7C14b77578977342d58507251ca2dc2b06%7C0%7C0%7C638539160969551214%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C0%7C%7C%7C&sdata=oGmBC7t0qV%2FXTkZQkvJttt5x2yqAQOwuisazqzYsfIU%3D&reserved=0<https://aka.ms/o0ukef>>
________________________________
From: ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu>
Sent: Thursday, June 13, 2024 6:30:53 PM
To: Browne, Kristen (NIH/NIAID) [C] <kristen.browne@nih.gov>;
pett@cgl.ucsf.edu <pett@cgl.ucsf.edu>
Cc: goddard@cgl.ucsf.edu <goddard@cgl.ucsf.edu>; Cruz, Phil (NIH/NIAID)
[C] <phil.cruz@nih.gov>
Subject: [EXTERNAL] Re: [ChimeraX] #15428: problem with viruses bychain
outputs in flows
#15428: problem with viruses bychain outputs in flows
---------------------------------------+----------------------
Reporter: kristen.browne@… | Owner: pett
Type: defect | Status: accepted
Priority: normal | Milestone:
Component: Depiction | Version:
Resolution: | Keywords:
Blocked By: | Blocking:
Notify when closed: | Platform: all
Project: ChimeraX |
---------------------------------------+----------------------
Changes (by pett):
* cc: Tom Goddard, phil.cruz@… (added)
* component: Unassigned => Depiction
* owner: (none) => pett
* platform: => all
* project: => ChimeraX
* status: new => accepted
Comment:
Hi Kristen,
The issue is that the pipeline script opens the structure with
"open pdbe_bio:6wdt maxassemblies 1", which produces chain IDs of A, AA,
AAA, through ABG for the 60 copies of chain A from the original
asymmetric
unit. Similarly for chains B-F of the asymmetric unit. Whereas if you
simply open 6wdt and click on the first entry of its assembly table, you
get 60 chain As, 60 chain Bs, etc. These 2 sets of chain IDs color
differently when you "rainbow chains" on them (which is what the preset
is
doing).
If you look at the pipeline "color bypolymer" output for 6wdt you
will see that the color arrangement is the same as the "by chain" you did
by hand (though the colors themselves will be different).
So the question is what do we want to do about this. There seems
to be metadata in the pdbe_bio file indicating that it is an assembly, so
it's possible that the by-chains preset could do something
different/smarter if it detects that.
--Eric
--
Ticket URL:
<https://gcc02.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.rbvi.ucsf.edu%2Ftrac%2FChimeraX%2Fticket%2F15428%23comment%3A1&data=05%7C02%7Ckristen.browne%40nih.gov%7C22ff71f196094e8e42c008dc8bfbd683%7C14b77578977342d58507251ca2dc2b06%7C0%7C0%7C638539160969560905%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C0%7C%7C%7C&sdata=wvBvol2JdWaTMa%2BZBGo2%2F73VHG6aPnE5%2BXhloR61YjY%3D&reserved=0<https://gcc02.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.rbvi.ucsf.edu%2Ftrac%2FChimeraX%2Fticket%2F15428%23comment%3A1&data=05%7C02%7Ckristen.browne%40nih.gov%7C22ff71f196094e8e42c008dc8bfbd683%7C14b77578977342d58507251ca2dc2b06%7C0%7C0%7C638539160969567550%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C0%7C%7C%7C&sdata=bc5ZKhHc%2BTc2EXDCprwgF7T%2BMXPjzPu6em8SzWqG8I8%3D&reserved=0<https://www.rbvi.ucsf.edu/trac/ChimeraX/ticket/15428#comment:1>>>
ChimeraX
<https://gcc02.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.rbvi.ucsf.edu%2Fchimerax%2F&data=05%7C02%7Ckristen.browne%40nih.gov%7C22ff71f196094e8e42c008dc8bfbd683%7C14b77578977342d58507251ca2dc2b06%7C0%7C0%7C638539160969571786%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C0%7C%7C%7C&sdata=Vi8vqyynw8PjvAa8E9%2Ffll6%2FKP2dtR6CuPVl17MET%2Bs%3D&reserved=0<https://gcc02.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.rbvi.ucsf.edu%2Fchimerax%2F&data=05%7C02%7Ckristen.browne%40nih.gov%7C22ff71f196094e8e42c008dc8bfbd683%7C14b77578977342d58507251ca2dc2b06%7C0%7C0%7C638539160969575831%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C0%7C%7C%7C&sdata=9PH%2FjwFDLQBQmU8gimyq0zX%2FOFtPRdk%2BWstZGcijPq0%3D&reserved=0<https://www.rbvi.ucsf.edu/chimerax/>>>
ChimeraX Issue Tracker
CAUTION: This email originated from outside of the organization. Do not
click links or open attachments unless you recognize the sender and are
confident the content is safe.
}}}
--
Ticket URL: <https://gcc02.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.rbvi.ucsf.edu%2Ftrac%2FChimeraX%2Fticket%2F15428%23comment%3A2&data=05%7C02%7Ckristen.browne%40nih.gov%7C22ff71f196094e8e42c008dc8bfbd683%7C14b77578977342d58507251ca2dc2b06%7C0%7C0%7C638539160969579822%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C0%7C%7C%7C&sdata=JAyBPKTSbC9N5zQNf6zHZmzvL4A7IGvNRwOJ1QNIOFQ%3D&reserved=0<https://www.rbvi.ucsf.edu/trac/ChimeraX/ticket/15428#comment:2>>
ChimeraX <https://gcc02.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.rbvi.ucsf.edu%2Fchimerax%2F&data=05%7C02%7Ckristen.browne%40nih.gov%7C22ff71f196094e8e42c008dc8bfbd683%7C14b77578977342d58507251ca2dc2b06%7C0%7C0%7C638539160969584173%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C0%7C%7C%7C&sdata=O%2FEEqmS111BpNfWJq71bRS%2FXgfjl0Kl4tfsdozGqOK0%3D&reserved=0<https://www.rbvi.ucsf.edu/chimerax/>>
ChimeraX Issue Tracker
CAUTION: This email originated from outside of the organization. Do not click links or open attachments unless you recognize the sender and are confident the content is safe.
comment:4 by , 21 months ago
| Cc: | added |
|---|
comment:5 by , 21 months ago
I have updated the presets bundle to look at the PDBE mmCIF table that maps final chain IDs to the original asymmetric unit IDs. I'll send an updated version of the bundle once the daily build is working again, since the change depends on a fix that will be in the next daily build.
comment:6 by , 21 months ago
| Resolution: | → fixed |
|---|---|
| Status: | accepted → closed |
The updated bundle is on the toolshed, and the pipeline script has been updated.
Added by email2trac