#15360 closed defect (duplicate)
Problems with the display of cyclic peptides
| Reported by: | Owned by: | Tom Goddard | |
|---|---|---|---|
| Priority: | normal | Milestone: | |
| Component: | Depiction | Version: | |
| Keywords: | Cc: | Eric Pettersen | |
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
The following bug report has been submitted:
Platform: macOS-12.0.1-arm64-arm-64bit
ChimeraX Version: 1.7.1 (2024-01-23 01:58:08 UTC)
Description
When drawing cartoon for a cyclic peptide the "termini" are drawn incorrectly. According to an earlier ticket this is a known issue. Is there any chance this could be fixed? We are submitting a paper where we are using Chimera for all figures and while using the shape tube option works well for peptides without secondary structure it becomes a bit unwieldy if there is a helix or sheet portion.
Thank you very much.
Log:
UCSF ChimeraX version: 1.7.1 (2024-01-23)
© 2016-2023 Regents of the University of California. All rights reserved.
> open /Users/stephenrettie/Desktop/mcb_d2_alignzoom.cxs
Log from Thu May 16 12:45:06 2024UCSF ChimeraX version: 1.7.1 (2024-01-23)
© 2016-2023 Regents of the University of California. All rights reserved.
> open /Users/stephenrettie/Desktop/mcl1_align.cxs
Log from Fri May 3 14:10:47 2024UCSF ChimeraX version: 1.7.1 (2024-01-23)
© 2016-2023 Regents of the University of California. All rights reserved.
> open /Users/stephenrettie/Desktop/mcl1_surface.cxs
Log from Fri May 3 13:53:46 2024UCSF ChimeraX version: 1.7.1 (2024-01-23)
© 2016-2023 Regents of the University of California. All rights reserved.
> open /Users/stephenrettie/Desktop/MCB_D2_figures_fixed_termini.cxs
Log from Tue Apr 30 14:54:36 2024UCSF ChimeraX version: 1.7.1 (2024-01-23)
© 2016-2023 Regents of the University of California. All rights reserved.
> open /Users/stephenrettie/Downloads/MCB_D2_figures_sidechains.cxs
Log from Fri Apr 26 16:37:40 2024UCSF ChimeraX version: 1.7.1 (2024-01-23)
© 2016-2023 Regents of the University of California. All rights reserved.
> open /Users/gauravbhardwaj/MCB_D2_figures.cxs
Log from Fri Apr 26 16:32:15 2024 Startup Messages
---
warning | Replacing fetcher for 'pdb_nmr' and format NMRSTAR from NMRSTAR
bundle with that from NMRSTAR bundle
UCSF ChimeraX version: 1.7.1 (2024-01-23)
© 2016-2023 Regents of the University of California. All rights reserved.
How to cite UCSF ChimeraX
> open /Users/gauravbhardwaj/Downloads/MCB_D2_AF2.pdb
Chain information for MCB_D2_AF2.pdb #1
---
Chain | Description
A | No description available
B | No description available
> open "/Users/gauravbhardwaj/Downloads/MCL1_2_refine_016 (1).pdb"
Chain information for MCL1_2_refine_016 (1).pdb #2
---
Chain | Description
A | No description available
B | No description available
> matchmaker #2 to #1
Parameters
---
Chain pairing | bb
Alignment algorithm | Needleman-Wunsch
Similarity matrix | BLOSUM-62
SS fraction | 0.3
Gap open (HH/SS/other) | 18/18/6
Gap extend | 1
SS matrix | | | H | S | O
---|---|---|---
H | 6 | -9 | -6
S | | 6 | -6
O | | | 4
Iteration cutoff | 2
Matchmaker MCB_D2_AF2.pdb, chain A (#1) with MCL1_2_refine_016 (1).pdb, chain
A (#2), sequence alignment score = 784.7
RMSD between 145 pruned atom pairs is 0.570 angstroms; (across all 149 pairs:
0.724)
> set bgColor white
> lighting soft
> graphics silhouettes true width 5
> color /A rgn(200,200,200)
Expected a color or one of 'byatom', 'bychain', 'byelement', 'byhetero',
'byidentity', 'bymodel', 'bynucleotide', 'bypolymer', 'fromatoms',
'fromcartoons', 'fromribbons', or 'random' or a keyword
> color /A rgb(200,200,200)
> hide #2 models
> show #2 models
> bond /B:1@N/B:16@C reasonable false
Created 1 bond
> cartoon /B suppressBackboneDisplay false
> show /B:1@N,CA/B:16@CA,C bonds
> size stickRadius 0.3
Changed 2701 bond radii
> hide #2 models
> show #2 models
> hide #1 models
> show #2/B:1@N,CA/#2B:16@CA,C bonds
Expected a collection of one of 'atoms', 'bonds', 'cartoons', 'models',
'pbonds', 'pseudobonds', 'ribbons', or 'surfaces' or a keyword
> bond #2/B:1@N#2/B:16@C reasonable false
Must specify two or more atoms
> bond #2/B:152@N#2/B:167@C reasonable false
Created 0 bonds
> show /B:152@N,CA/B:1667@CA,C bonds
> show /B:152@N,CA/B:167@CA,C bonds
> artoon style width 2 thick 0.6
Unknown command: artoon style width 2 thick 0.6
> cartoon tether /B shape cone sides 4 scale 0 opacity 0
[Repeated 1 time(s)]
> cartoon style width 2 thickness 0.6
> color name coco_red rgb(255,0,0)
Color 'coco_red' is opaque: rgb(100%, 0%, 0%) hex: #ff0000
> color name coco_blue rgb(0,157,246)
Color 'coco_blue' is opaque: rgb(0%, 61.6%, 96.5%) hex: #009df6
> color name coco_orange rgb(255,156,0)
Color 'coco_orange' is opaque: rgb(100%, 61.2%, 0%) hex: #ff9c00
> color name coco_magenta rgb(255,0,148)
Color 'coco_magenta' is opaque: rgb(100%, 0%, 58%) hex: #ff0094
> color name coco_magenta2 rgb(206,0,119)
Color 'coco_magenta2' is opaque: rgb(80.8%, 0%, 46.7%) hex: #ce0077
> color name xkcd_green rgb(21,176,26)
Color 'xkcd_green' is opaque: rgb(8.24%, 69%, 10.2%) hex: #15b01a
> color name xkcd_pink rgb(255,129,192)
Color 'xkcd_pink' is opaque: rgb(100%, 50.6%, 75.3%) hex: #ff81c0
> color name xkcd_violet rgb(154,14,234)
Color 'xkcd_violet' is opaque: rgb(60.4%, 5.49%, 91.8%) hex: #9a0eea
> color name xkcd_periwinkle rgb(142,130,254)
Color 'xkcd_periwinkle' is opaque: rgb(55.7%, 51%, 99.6%) hex: #8e82fe
> color name xkcd_gold rgb(219,180,12)
Color 'xkcd_gold' is opaque: rgb(85.9%, 70.6%, 4.71%) hex: #dbb40c
> color name xkcd_bluegreen rgb(19,126,109)
Color 'xkcd_bluegreen' is opaque: rgb(7.45%, 49.4%, 42.7%) hex: #137e6d
> color name xkcd_cerulean rgb(4,133,209)
Color 'xkcd_cerulean' is opaque: rgb(1.57%, 52.2%, 82%) hex: #0485d1
> color name xkcd_marigold rgb(252,192,6)
Color 'xkcd_marigold' is opaque: rgb(98.8%, 75.3%, 2.35%) hex: #fcc006
> color name tomato2 rgb(245,96,42)
Color 'tomato2' is opaque: rgb(96.1%, 37.6%, 16.5%) hex: #f5602a
> color name hblue rgb(54,175,178)
Color 'hblue' is opaque: rgb(21.2%, 68.6%, 69.8%) hex: #36afb2
> color name hgreen rgb(139,194,70)
Color 'hgreen' is opaque: rgb(54.5%, 76.1%, 27.5%) hex: #8bc246
> color name hblue2 rgb(83,155,186)
Color 'hblue2' is opaque: rgb(32.5%, 60.8%, 72.9%) hex: #539bba
> color name light_red rgb(249,128,111)
Color 'light_red' is opaque: rgb(97.6%, 50.2%, 43.5%) hex: #f9806f
> color name yelloworange rgb(250,181,96)
Color 'yelloworange' is opaque: rgb(98%, 71%, 37.6%) hex: #fab560
> color name brightyellow rgb(255,202,3)
Color 'brightyellow' is opaque: rgb(100%, 79.2%, 1.18%) hex: #ffca03
> color name new_gray rgb(200,200,200)
Color 'new_gray' is opaque: gray(78.4%) hex: #c8c8c8
> color #/B xkcd_marigold
Expected a color or one of 'byatom', 'bychain', 'byelement', 'byhetero',
'byidentity', 'bymodel', 'bynucleotide', 'bypolymer', 'fromatoms',
'fromcartoons', 'fromribbons', or 'random' or a keyword
> color #2/B xkcd_marigold
> color #1/B coco_blue
> show #1 models
> color #1/B cotnflower blue
Expected a color or one of 'byatom', 'bychain', 'byelement', 'byhetero',
'byidentity', 'bymodel', 'bynucleotide', 'bypolymer', 'fromatoms',
'fromcartoons', 'fromribbons', or 'random' or a keyword
> color #1/B cornflower blue
> color #1/B xkcd_cerulean
> hide #2/A cartoons
> save /Users/gauravbhardwaj/mcb_D2_model_xray_chimeraX.png supersample 10
> color #1/B xkcd_violet
> color #1/B xkcd_periwinkle
> color #1/B xkcd_violet
> color #1/B coco_blue
> color #1/B coco_orange
> color #1/B coco_blue
> color #2/B coco_oramge
Expected a color or one of 'byatom', 'bychain', 'byelement', 'byhetero',
'byidentity', 'bymodel', 'bynucleotide', 'bypolymer', 'fromatoms',
'fromcartoons', 'fromribbons', or 'random' or a keyword
> color #2/B coco_orange
> color #2/B xkcd_marigold
> color #2/B xkcd_violet
> color #1/B xkcd_violet
> save /Users/gauravbhardwaj/mcb_D2_model_xray_chimeraX.png supersample 10
> hide #2 models
> show #1/a surfaces
> show #1/b and sidechain bonds
Expected a collection of one of 'atoms', 'bonds', 'cartoons', 'models',
'pbonds', 'pseudobonds', 'ribbons', or 'surfaces' or a keyword
> show #1/a & sidechain bonds
> show #1/a & sidechain atoms
> show #1/b & sidechain atoms
> show #1/b:2@N atoms
> color #1 byhetero
> color #1/b byhetero
> hide #1/a atoms
> hide #1/a bonds
> save /Users/gauravbhardwaj/mcb_D2_model_surface_chimeraX.png supersample 10
> save /Users/gauravbhardwaj/mcb_D2_model_surface_no_sticks_chimeraX.png
> supersample 10
> save /Users/gauravbhardwaj/MCB_D2_figures.cxs
——— End of log from Fri Apr 26 16:32:15 2024 ———
opened ChimeraX session
> hide #1/a surfaces
> hide #1/a bonds
> hide #1/a stoms
Expected a collection of one of 'atoms', 'bonds', 'cartoons', 'models',
'pbonds', 'pseudobonds', 'ribbons', or 'surfaces' or a keyword
> hide #1/a atoms
> show #2 models
> hide /b atoms
> show /b atoms
> show /b and sidechain atoms
Expected a collection of one of 'atoms', 'bonds', 'cartoons', 'models',
'pbonds', 'pseudobonds', 'ribbons', or 'surfaces' or a keyword
> show /b & sidechain atoms
> show #1/b:2@N atoms
> show #2/b:153@N atoms
> color /b byhetero
> save /Users/gauravbhardwaj/MCB_D2_figures_sidechains.cxs
——— End of log from Fri Apr 26 16:37:40 2024 ———
opened ChimeraX session
> select #1/B:1@CA
1 atom, 1 residue, 1 model selected
> hide sel cartoons
> select #2/B:152
7 atoms, 7 bonds, 1 residue, 1 model selected
> hide sel cartoons
[Repeated 1 time(s)]
> select #1/B:16
5 atoms, 4 bonds, 1 residue, 1 model selected
> hide sel cartoons
> shape tube #2/B@ca radius 0.3
> hide #!1 target m
> hide #3 target m
> shape tube #2/B:152-162@ca radius 0.3
> close #4
> shape tube #2/B:162-152@ca radius 0.3
No atoms specified
> shape tube #2/B:166-167@ca radius 0.3
> shape tube #2/B:162-167@ca radius 0.3
> close #4
> show sel cartoons
> select #5
1 model selected
> show #3 target m
> close #3
> hide #5 target m
> select #2/B:166
8 atoms, 7 bonds, 1 residue, 1 model selected
> hide sel cartoons
> show #5 target m
> color tube gold
Expected a color or one of 'byatom', 'bychain', 'byelement', 'byhetero',
'byidentity', 'bymodel', 'bynucleotide', 'bypolymer', 'fromatoms',
'fromcartoons', 'fromribbons', or 'random' or a keyword
> color #3 gold
> color #5 gold
> select #2/B:165
9 atoms, 8 bonds, 1 residue, 1 model selected
> hide sel cartoons
> select #2/B:164
8 atoms, 7 bonds, 1 residue, 1 model selected
> hide sel cartoons
> select #2/B:163
4 atoms, 3 bonds, 1 residue, 1 model selected
> hide sel cartoons
> select #2/B:162
7 atoms, 6 bonds, 1 residue, 1 model selected
> hide sel cartoons
> show sel cartoons
> select clear
> select #5
1 model selected
> color #5 rgb(219,180,12)
> select #2/B:157
14 atoms, 15 bonds, 1 residue, 1 model selected
> color #5 rgb(252,192,6)
> select #5
1 model selected
> hide #5 target m
> select #2/B:163@CA
1 atom, 1 residue, 1 model selected
> show sel cartoons
> show #5 models
> hide #5 models
> select #2/B:164@CA
1 atom, 1 residue, 1 model selected
> show sel cartoons
> show #5 models
> show #1/B:152@c
> hide #1/B:152@c
> show #2/B:152@c
> hide pseudobonds
> select #2/B:153
7 atoms, 7 bonds, 1 residue, 1 model selected
> hide sel cartoons
> show sel cartoons
> show #!1 models
> hide #!2 models
> hide #5 models
> shape tube #1/B:1-12@ca radius 0.3
> hide #5 models
> close #3
> shape tube #1/B:11-16@ca radius 0.3
> select #1/A:17
9 atoms, 8 bonds, 1 residue, 1 model selected
> select #1/B:1@CA
1 atom, 1 residue, 1 model selected
> show sel cartoons
> hide sel cartoons
> show #1/B:1@c
> show #!2 models
> hide #!2 models
> color #3 rgb(154,14,234)
> save MCB_D2_figures_fixed_termini.csx
No known data format for file suffix '.csx'
> save MCB_D2_figures_fixed_termini.cxs
——— End of log from Tue Apr 30 14:54:36 2024 ———
opened ChimeraX session
> select #2/A
1202 atoms, 1222 bonds, 150 residues, 1 model selected
> select #1/A
1217 atoms, 1237 bonds, 151 residues, 1 model selected
> show sel surfaces
> hide #1/B:3 * sidechain
Expected a collection of one of 'atoms', 'bonds', 'cartoons', 'models',
'pbonds', 'pseudobonds', 'ribbons', or 'surfaces' or a keyword
> hide #1/B:3 & sidechain
> show #1/B:3@ca
> hide #1/B:3@ca
> hide #1/B:9 & sidechain
> undo
> hide #1/B:9 & sidechain
> hide #1/B:5 & sidechain
> hide #1/B:14 & sidechain
> hide #1/B:15 & sidechain
> ~select
Nothing selected
> hide #1/B:2 & sidechain
> show #1/B:2@ca
> select #1/B:2@N
1 atom, 1 residue, 1 model selected
> select #1/B:2@N
1 atom, 1 residue, 1 model selected
> select #1/B:2@N
1 atom, 1 residue, 1 model selected
> color #1/B:2@n rgb(154,14,234)
> ~select
Nothing selected
> save mcl1_surface.cxs
——— End of log from Fri May 3 13:53:46 2024 ———
opened ChimeraX session
> save mcl1_surface.png supersample 20 transparentBackground true
> show #!2 models
> hide /B & sidechain
> color #1/B rgb(154,14,234)
> color #2/B rgb(252,192,6)
> show /B:2@ca
> hide #2/B:2@ca
> show /B:153@ca
> show /B:167@ca
> show /B:167@c
> show #5 models
> show #1/B:1@ca
> show #1/B:2@ca
> show #1/B:2@n
> show #1/B:16@c
[Repeated 1 time(s)]
> show #1/B:16@ca
> show #2/B:152@ca
> select /A
2419 atoms, 2459 bonds, 301 residues, 2 models selected
> hide sel surfaces
> ~select
Nothing selected
> save mcl1_align.cxs
——— End of log from Fri May 3 14:10:47 2024 ———
opened ChimeraX session
> show cartoons #2/A
Expected ',' or a keyword
> show #2/A cartoons
> hide #1/A cartoons
> save mcbd2_xtal_align.png supersample 20 transparentBackground true
[Repeated 1 time(s)]
> show /B sidechain
Expected a collection of one of 'atoms', 'bonds', 'cartoons', 'models',
'pbonds', 'pseudobonds', 'ribbons', or 'surfaces' or a keyword
> show /B & sidechain
> hide pbonds
> color sidechain byhetero
> color /B:pro@n byhetero
> save mcbd2_xtal_align_zoom.png supersample 20 transparentBackground true
> save mcb_d2_alignzoom.cxs
——— End of log from Thu May 16 12:45:06 2024 ———
opened ChimeraX session
> open
> /Users/stephenrettie/Downloads/fold_2024_05_31_16_16/fold_2024_05_31_16_16_model_0.cif
Chain information for fold_2024_05_31_16_16_model_0.cif #4
---
Chain | Description
A | .
B | .
OpenGL version: 4.1 Metal - 76.1
OpenGL renderer: Apple M1
OpenGL vendor: Apple
Python: 3.11.2
Locale: UTF-8
Qt version: PyQt6 6.3.1, Qt 6.3.1
Qt runtime version: 6.3.2
Qt platform: cocoa
Hardware:
Hardware Overview:
Model Name: MacBook Pro
Model Identifier: MacBookPro17,1
Chip: Apple M1
Total Number of Cores: 8 (4 performance and 4 efficiency)
Memory: 8 GB
System Firmware Version: 7429.41.5
OS Loader Version: 7429.41.5
Software:
System Software Overview:
System Version: macOS 12.0.1 (21A559)
Kernel Version: Darwin 21.1.0
Time since boot: 10 days 17:24
Graphics/Displays:
Apple M1:
Chipset Model: Apple M1
Type: GPU
Bus: Built-In
Total Number of Cores: 8
Vendor: Apple (0x106b)
Metal Family: Supported, Metal GPUFamily Apple 7
Displays:
Color LCD:
Display Type: Built-In Retina LCD
Resolution: 2560 x 1600 Retina
Main Display: Yes
Mirror: Off
Online: Yes
Automatically Adjust Brightness: No
Connection Type: Internal
BenQ PD2700U:
Resolution: 3840 x 2160 (2160p/4K UHD 1 - Ultra High Definition)
UI Looks like: 1920 x 1080 @ 30.00Hz
Mirror: Off
Online: Yes
Rotation: Supported
Automatically Adjust Brightness: Yes
Installed Packages:
alabaster: 0.7.16
appdirs: 1.4.4
appnope: 0.1.3
asttokens: 2.4.1
Babel: 2.14.0
backcall: 0.2.0
beautifulsoup4: 4.11.2
blockdiag: 3.0.0
blosc2: 2.0.0
build: 0.10.0
certifi: 2022.12.7
cftime: 1.6.3
charset-normalizer: 3.3.2
ChimeraX-AddCharge: 1.5.13
ChimeraX-AddH: 2.2.5
ChimeraX-AlignmentAlgorithms: 2.0.1
ChimeraX-AlignmentHdrs: 3.4.1
ChimeraX-AlignmentMatrices: 2.1
ChimeraX-Alignments: 2.12.2
ChimeraX-AlphaFold: 1.0
ChimeraX-AltlocExplorer: 1.1.1
ChimeraX-AmberInfo: 1.0
ChimeraX-Arrays: 1.1
ChimeraX-Atomic: 1.49.1
ChimeraX-AtomicLibrary: 12.1.5
ChimeraX-AtomSearch: 2.0.1
ChimeraX-AxesPlanes: 2.3.2
ChimeraX-BasicActions: 1.1.2
ChimeraX-BILD: 1.0
ChimeraX-BlastProtein: 2.1.2
ChimeraX-BondRot: 2.0.4
ChimeraX-BugReporter: 1.0.1
ChimeraX-BuildStructure: 2.10.5
ChimeraX-Bumps: 1.0
ChimeraX-BundleBuilder: 1.2.2
ChimeraX-ButtonPanel: 1.0.1
ChimeraX-CageBuilder: 1.0.1
ChimeraX-CellPack: 1.0
ChimeraX-Centroids: 1.3.2
ChimeraX-ChangeChains: 1.1
ChimeraX-CheckWaters: 1.3.2
ChimeraX-ChemGroup: 2.0.1
ChimeraX-Clashes: 2.2.4
ChimeraX-ColorActions: 1.0.3
ChimeraX-ColorGlobe: 1.0
ChimeraX-ColorKey: 1.5.5
ChimeraX-CommandLine: 1.2.5
ChimeraX-ConnectStructure: 2.0.1
ChimeraX-Contacts: 1.0.1
ChimeraX-Core: 1.7.1
ChimeraX-CoreFormats: 1.2
ChimeraX-coulombic: 1.4.2
ChimeraX-Crosslinks: 1.0
ChimeraX-Crystal: 1.0
ChimeraX-CrystalContacts: 1.0.1
ChimeraX-DataFormats: 1.2.3
ChimeraX-Dicom: 1.2
ChimeraX-DistMonitor: 1.4
ChimeraX-DockPrep: 1.1.3
ChimeraX-Dssp: 2.0
ChimeraX-EMDB-SFF: 1.0
ChimeraX-ESMFold: 1.0
ChimeraX-FileHistory: 1.0.1
ChimeraX-FunctionKey: 1.0.1
ChimeraX-Geometry: 1.3
ChimeraX-gltf: 1.0
ChimeraX-Graphics: 1.1.1
ChimeraX-Hbonds: 2.4
ChimeraX-Help: 1.2.2
ChimeraX-HKCage: 1.3
ChimeraX-IHM: 1.1
ChimeraX-ImageFormats: 1.2
ChimeraX-IMOD: 1.0
ChimeraX-IO: 1.0.1
ChimeraX-ItemsInspection: 1.0.1
ChimeraX-IUPAC: 1.0
ChimeraX-Label: 1.1.8
ChimeraX-ListInfo: 1.2.2
ChimeraX-Log: 1.1.6
ChimeraX-LookingGlass: 1.1
ChimeraX-Maestro: 1.9.1
ChimeraX-Map: 1.1.4
ChimeraX-MapData: 2.0
ChimeraX-MapEraser: 1.0.1
ChimeraX-MapFilter: 2.0.1
ChimeraX-MapFit: 2.0
ChimeraX-MapSeries: 2.1.1
ChimeraX-Markers: 1.0.1
ChimeraX-Mask: 1.0.2
ChimeraX-MatchMaker: 2.1.2
ChimeraX-MCopy: 1.0
ChimeraX-MDcrds: 2.6.1
ChimeraX-MedicalToolbar: 1.0.2
ChimeraX-Meeting: 1.0.1
ChimeraX-MLP: 1.1.1
ChimeraX-mmCIF: 2.12.1
ChimeraX-MMTF: 2.2
ChimeraX-Modeller: 1.5.14
ChimeraX-ModelPanel: 1.4
ChimeraX-ModelSeries: 1.0.1
ChimeraX-Mol2: 2.0.3
ChimeraX-Mole: 1.0
ChimeraX-Morph: 1.0.2
ChimeraX-MouseModes: 1.2
ChimeraX-Movie: 1.0
ChimeraX-Neuron: 1.0
ChimeraX-Nifti: 1.1
ChimeraX-NRRD: 1.1
ChimeraX-Nucleotides: 2.0.3
ChimeraX-OpenCommand: 1.13.1
ChimeraX-PDB: 2.7.3
ChimeraX-PDBBio: 1.0.1
ChimeraX-PDBLibrary: 1.0.4
ChimeraX-PDBMatrices: 1.0
ChimeraX-PickBlobs: 1.0.1
ChimeraX-Positions: 1.0
ChimeraX-PresetMgr: 1.1
ChimeraX-PubChem: 2.1
ChimeraX-ReadPbonds: 1.0.1
ChimeraX-Registration: 1.1.2
ChimeraX-RemoteControl: 1.0
ChimeraX-RenderByAttr: 1.1
ChimeraX-RenumberResidues: 1.1
ChimeraX-ResidueFit: 1.0.1
ChimeraX-RestServer: 1.2
ChimeraX-RNALayout: 1.0
ChimeraX-RotamerLibMgr: 4.0
ChimeraX-RotamerLibsDunbrack: 2.0
ChimeraX-RotamerLibsDynameomics: 2.0
ChimeraX-RotamerLibsRichardson: 2.0
ChimeraX-SaveCommand: 1.5.1
ChimeraX-SchemeMgr: 1.0
ChimeraX-SDF: 2.0.2
ChimeraX-Segger: 1.0
ChimeraX-Segment: 1.0.1
ChimeraX-SelInspector: 1.0
ChimeraX-SeqView: 2.11
ChimeraX-Shape: 1.0.1
ChimeraX-Shell: 1.0.1
ChimeraX-Shortcuts: 1.1.1
ChimeraX-ShowSequences: 1.0.2
ChimeraX-SideView: 1.0.1
ChimeraX-Smiles: 2.1.2
ChimeraX-SmoothLines: 1.0
ChimeraX-SpaceNavigator: 1.0
ChimeraX-StdCommands: 1.12.4
ChimeraX-STL: 1.0.1
ChimeraX-Storm: 1.0
ChimeraX-StructMeasure: 1.1.2
ChimeraX-Struts: 1.0.1
ChimeraX-Surface: 1.0.1
ChimeraX-SwapAA: 2.0.1
ChimeraX-SwapRes: 2.2.2
ChimeraX-TapeMeasure: 1.0
ChimeraX-TaskManager: 1.0
ChimeraX-Test: 1.0
ChimeraX-Toolbar: 1.1.2
ChimeraX-ToolshedUtils: 1.2.4
ChimeraX-Topography: 1.0
ChimeraX-ToQuest: 1.0
ChimeraX-Tug: 1.0.1
ChimeraX-UI: 1.33.3
ChimeraX-uniprot: 2.3
ChimeraX-UnitCell: 1.0.1
ChimeraX-ViewDockX: 1.3.2
ChimeraX-VIPERdb: 1.0
ChimeraX-Vive: 1.1
ChimeraX-VolumeMenu: 1.0.1
ChimeraX-vrml: 1.0
ChimeraX-VTK: 1.0
ChimeraX-WavefrontOBJ: 1.0
ChimeraX-WebCam: 1.0.2
ChimeraX-WebServices: 1.1.3
ChimeraX-Zone: 1.0.1
colorama: 0.4.6
comm: 0.2.1
contourpy: 1.2.0
cxservices: 1.2.2
cycler: 0.12.1
Cython: 0.29.33
debugpy: 1.8.0
decorator: 5.1.1
docutils: 0.19
executing: 2.0.1
filelock: 3.9.0
fonttools: 4.47.2
funcparserlib: 2.0.0a0
glfw: 2.6.4
grako: 3.16.5
h5py: 3.10.0
html2text: 2020.1.16
idna: 3.6
ihm: 0.38
imagecodecs: 2023.9.18
imagesize: 1.4.1
ipykernel: 6.23.2
ipython: 8.14.0
ipython-genutils: 0.2.0
ipywidgets: 8.1.1
jedi: 0.18.2
Jinja2: 3.1.2
jupyter-client: 8.2.0
jupyter-core: 5.7.1
jupyterlab-widgets: 3.0.9
kiwisolver: 1.4.5
line-profiler: 4.0.2
lxml: 4.9.2
lz4: 4.3.2
MarkupSafe: 2.1.4
matplotlib: 3.7.2
matplotlib-inline: 0.1.6
msgpack: 1.0.4
nest-asyncio: 1.6.0
netCDF4: 1.6.2
networkx: 3.1
nibabel: 5.0.1
nptyping: 2.5.0
numexpr: 2.8.8
numpy: 1.25.1
openvr: 1.23.701
packaging: 21.3
ParmEd: 3.4.3
parso: 0.8.3
pep517: 0.13.0
pexpect: 4.9.0
pickleshare: 0.7.5
pillow: 10.2.0
pip: 23.0
pkginfo: 1.9.6
platformdirs: 4.1.0
prompt-toolkit: 3.0.43
psutil: 5.9.5
ptyprocess: 0.7.0
pure-eval: 0.2.2
py-cpuinfo: 9.0.0
pycollada: 0.7.2
pydicom: 2.3.0
Pygments: 2.16.1
pynrrd: 1.0.0
PyOpenGL: 3.1.7
PyOpenGL-accelerate: 3.1.7
pyopenxr: 1.0.2801
pyparsing: 3.0.9
pyproject-hooks: 1.0.0
PyQt6-commercial: 6.3.1
PyQt6-Qt6: 6.3.2
PyQt6-sip: 13.4.0
PyQt6-WebEngine-commercial: 6.3.1
PyQt6-WebEngine-Qt6: 6.3.2
python-dateutil: 2.8.2
pytz: 2023.3.post1
pyzmq: 25.1.2
qtconsole: 5.4.3
QtPy: 2.4.1
RandomWords: 0.4.0
requests: 2.31.0
scipy: 1.11.1
setuptools: 67.4.0
setuptools-scm: 7.0.5
sfftk-rw: 0.7.3
six: 1.16.0
snowballstemmer: 2.2.0
sortedcontainers: 2.4.0
soupsieve: 2.5
sphinx: 6.1.3
sphinx-autodoc-typehints: 1.22
sphinxcontrib-applehelp: 1.0.8
sphinxcontrib-blockdiag: 3.0.0
sphinxcontrib-devhelp: 1.0.6
sphinxcontrib-htmlhelp: 2.0.5
sphinxcontrib-jsmath: 1.0.1
sphinxcontrib-qthelp: 1.0.7
sphinxcontrib-serializinghtml: 1.1.10
stack-data: 0.6.3
superqt: 0.5.0
tables: 3.8.0
tcia-utils: 1.5.1
tifffile: 2023.7.18
tinyarray: 1.2.4
tomli: 2.0.1
tornado: 6.4
traitlets: 5.9.0
typing-extensions: 4.9.0
tzdata: 2023.4
urllib3: 2.1.0
wcwidth: 0.2.13
webcolors: 1.12
wheel: 0.38.4
wheel-filename: 1.4.1
widgetsnbextension: 4.0.9
Attachments (3)
Change History (18)
comment:1 by , 21 months ago
| Component: | Unassigned → Depiction |
|---|---|
| Owner: | set to |
| Platform: | → all |
| Project: | → ChimeraX |
| Status: | new → assigned |
| Summary: | ChimeraX bug report submission → Problems with the display of cyclic peptides |
comment:2 by , 21 months ago
| Resolution: | → duplicate |
|---|---|
| Status: | assigned → closed |
Are you saying you have a structure with a cyclic peptide that contains a helix or sheet? I thought the cyclic peptides were typically short.
At any rate, closing the ends of a ribbon smoothly for cyclic peptides adds great complexity to the code for a rare case so I have not worked on it. I believe our old Chimera program (not ChimeraX) did it so you may want to use Chimera for making a circular peptide image.
comment:3 by , 21 months ago
Hello, Yes our manuscript contains two co-crystal structures and we have many designs that contain significant helical regions as well as sheets. The cartoon representation draws the cartoon beyond the Ca of the terminal residues which is very distracting, and turning cartoon off for the termini leaves a significant section missing. In PyMOL we usually just show the termini as sticks and things line up pretty well but that isn't as visually pleasing in ChimeraX. In our previous works we've used PyMOL renders, but really fell in love with the soft lighting and outlines in ChimeraX so we've moved over to it. As I said in the bug-report we've mostly gotten around this by showing non-secondary structure regions as tubes and then showing the terminal amide as atoms but it's not ideal and is kind of time consuming for each individual structure, and since each structure is different it's not straightforward to script. We are a design lab so we have lots of design models to show, not just the crystal structures. Thanks for your reply, I totally understand that our lab is one of very few labs for which this would be helpful. I'll see if I can get the same lighting settings in Chimera. On Tue, Jun 4, 2024 at 10:01 AM ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu> wrote: > > > > >
comment:4 by , 21 months ago
Could you attach a cyclic peptide structure of the type you want to show so I can play with it a bit? There are probably hacks that would make it join, like duplicating residues at the join and hiding those, but they are probably too much trouble for you if you want to do it frequently. But maybe with an example structure I will come up with some other trick which gets you closer to what you want to see.
comment:5 by , 21 months ago
I appreciate the offer, thank you! On Tue, Jun 4, 2024 at 2:59 PM ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu> wrote: > >
comment:6 by , 21 months ago
The attached PDB file is not cyclic. I think it would need a connect record to add the bond that joins the the first and last residues. Could you provide a file that properly defines the cyclic peptide?
comment:7 by , 21 months ago
Of course I can add the bond. But if the point is to find a way for ChimeraX to show ribbons as closed it really needs to have a correct file that says the loop is closed.
comment:8 by , 21 months ago
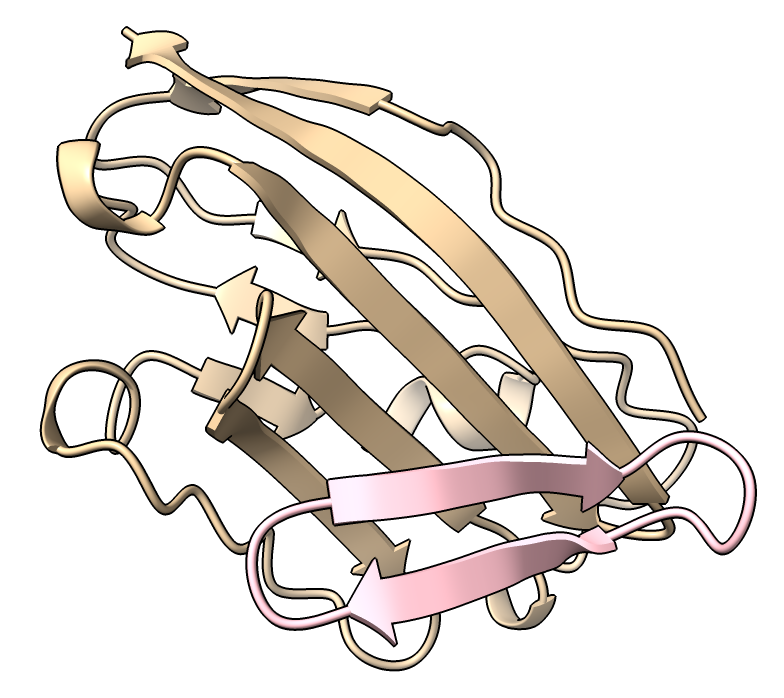
Cancel that request for the cyclic PDB file. It is easier to make the cyclic closed ribbon without the bond. Here I attach an image of the ribbon looking perfectly closed and the ChimeraX commands to do it. It is a bit cumbersome taking 6 commands to append a copy of the circular peptide to itself and then just show the middle part of the double length (two full cycle) peptide. The hidden ends of the longer peptide makes the ribbon close perfectly. This is fine for making a few publication images, but tedious if you want to do it for lots of structures. If you wanted to do it for more than 10 structures it would probably be worth writing a bit of Python that defines a single command that does it. The Python would just execute the same commands but would be needed to handle peptides of varying lengths.
comment:9 by , 21 months ago
Thank you!!! Can easily write a bit of python to make this easy but just knowing it works is massive! Thank you so much for your time spent on this. Have a great rest if your week. On Tue, Jun 4, 2024 at 6:44 PM ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu> wrote: > > > > >
comment:11 by , 21 months ago
Would be good to not have that trick buried in a Trac ticket -- I don't think tickets get indexed by Google. I can make a chimerax-recipes page for it today. On the other hand very few people work with cyclic peptides so I'm not sure I would spam chimerax-users. Tweeting would be more likely to reach the few people who deal in cyclic peptides.
comment:12 by , 21 months ago
I added a ChimeraX recipe web page showing this trick.
https://rbvi.github.io/chimerax-recipes/cyclic_peptide/cyclic.html
comment:13 by , 21 months ago
I keep getting the error 'Expected a keyword' when using the retainIds option. This is just me running the script you sent, it highlights retainIds for the error. I'm using 1.7.1. On Wed, Jun 5, 2024 at 1:14 PM ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu> wrote: > > >
comment:14 by , 21 months ago
The "retainIds" option to "combine" was added just after the 1.7.1 release, so you would need to use the 1.8 release candidate in order to get this to work.
comment:15 by , 21 months ago
Great thanks! On Thu, Jun 6, 2024, 3:01 PM ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu> wrote: > >
Reported by Stephen Rettie.
Adding him to recipient list for #7140