Opened 2 years ago
Closed 2 years ago
#14880 closed enhancement (fixed)
defattr Attribute File recipient value molecules or structures
| Reported by: | Owned by: | Eric Pettersen | |
|---|---|---|---|
| Priority: | normal | Milestone: | |
| Component: | Structure Editing | Version: | |
| Keywords: | Cc: | Elaine Meng | |
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
Hello, I am having difficulty with my *.defattr attribute file, and I am interested to know if there was a change in the value of recipient from "molecules" to "structures" at some point? I was using ChimeraX on two different machines, and one instance of ChimeraX refers to "molecules" and the other refers to "structures". Best, Chris Caffalette
Attachments (4)
Change History (8)
comment:2 by , 2 years ago
| Component: | Unassigned → Structure Editing |
|---|---|
| Owner: | set to |
| Platform: | → all |
| Project: | → ChimeraX |
| Status: | new → accepted |
| Type: | defect → enhancement |
Hi Chris,
The defattr file uses "molecules" to be backwards compatible with files used with our previous Chimera program. With ChimeraX we prefer the nomenclature "structures", which is why you see that in the Render By Attribute interface. It would seem good to allow that same term in defattr files (along with backwards-compatible "molecules").
Though the setattr command allows setting chain attributes, the defattr file doesn't currently support it. It probably should. We don't currently show chain attributes in the Render By Attribute tool basically because there are no interesting chain attributes to render by. If there was some compelling use case, that could be changed.
--Eric
Eric Pettersen
UCSF Computer Graphics Lab
comment:3 by , 2 years ago
Hi Eric,
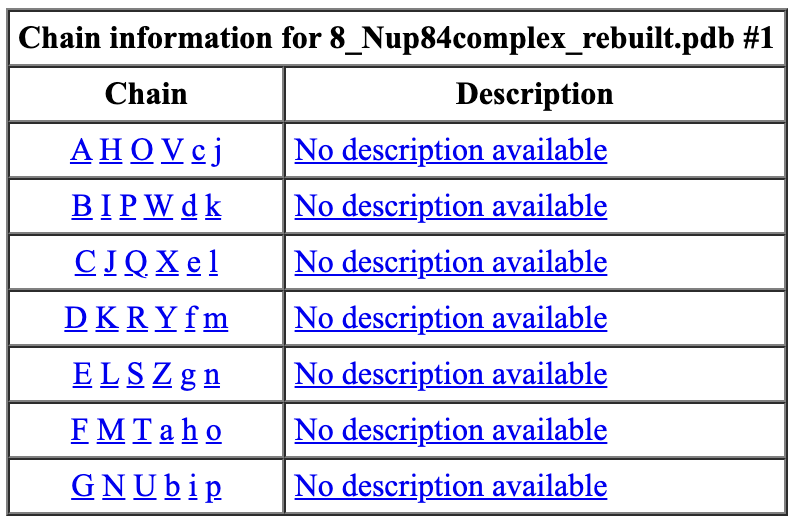
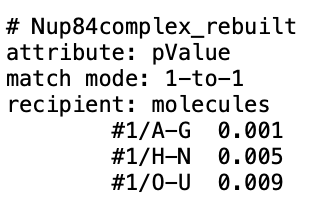
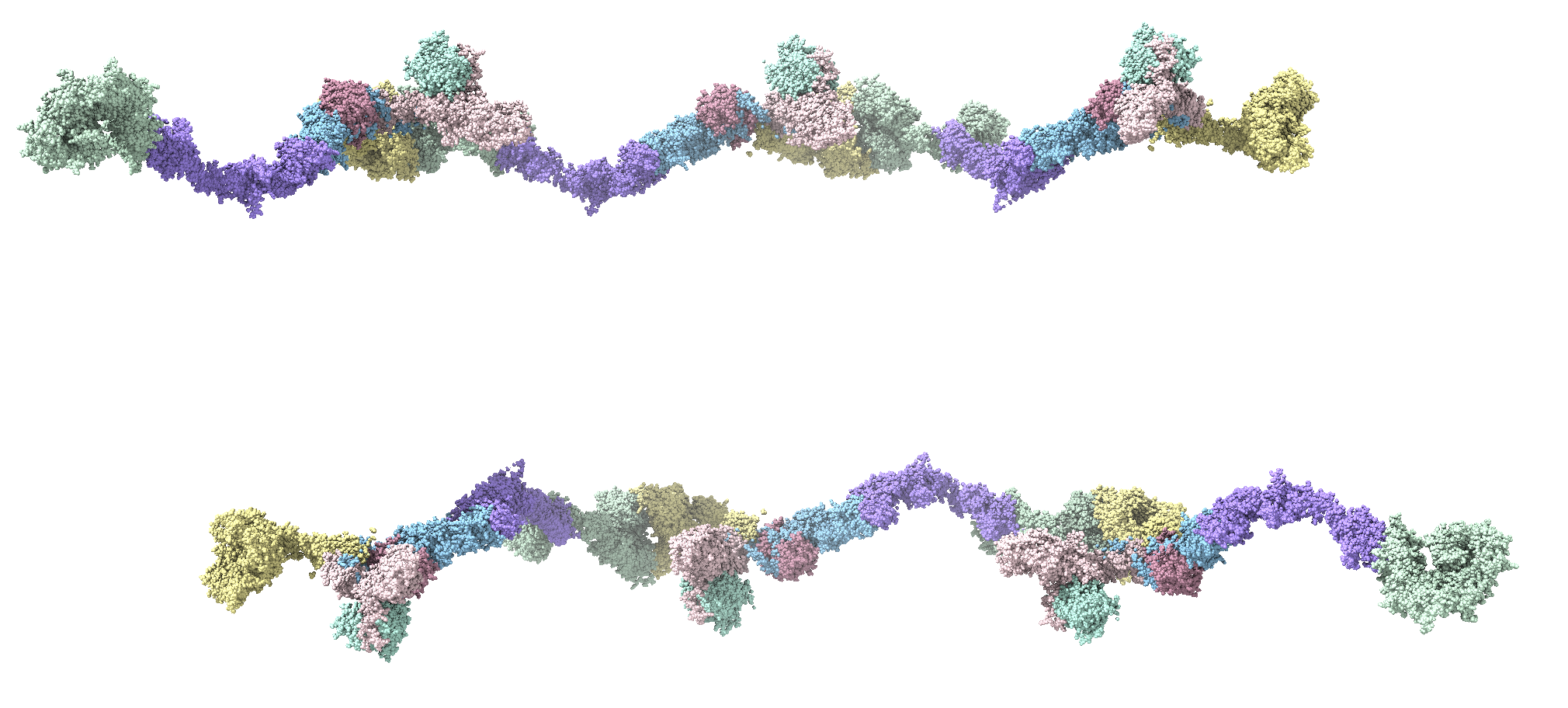
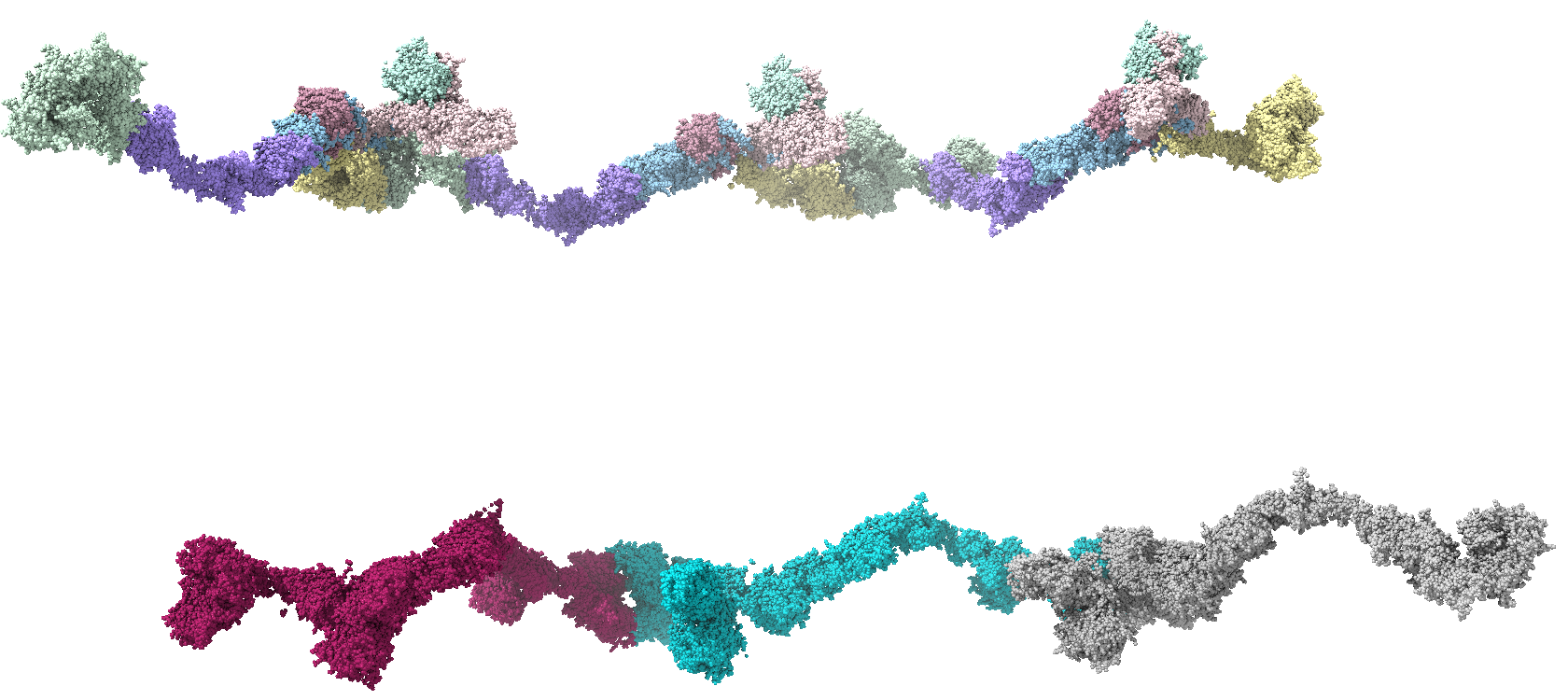
Thank you so much for this information. All of this stems from the desire to create a custom attribute, such as "pValue", and visualize an atomic model with a heat-map-like coloring of chains according to experimentally-determined numbers. In this particular case, the atomic models are of large complexes with many Chains representing the individual proteins of the complex. I originally, thought that I could use the atom-spec nomenclature to specify a range of Chains for molecule ID #1, such as #1/A-F, in a *.defattr attribute assignment file. However, since you mentioned that the setattr command will accept chains, I will instead create a complex setattr command delimited by semicolons to perform the same assignment of a custom attribute with "create true" e.g. the following setattr command and the cyan-grey-maroon palette in Render by Attribute.
setattr #1/A-G res pValue 0.001 create true; setattr #1/H-N res pValue 0.005 create true; setattr #1/O-U res pValue 0.009 create true
[cid:5e3d89e7-4222-4442-b903-0232a233b87b]
[cid:90075dcc-aac8-4398-a526-98a0f50da491][cid:0a263e08-bade-4f4c-8995-67750a89308e]
[cid:54b17d82-f808-490a-a7e4-30f50587b0a3]
Thank you again, Eric!
Best,
Chris Caffalette
________________________________
From: ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu>
Sent: Wednesday, April 3, 2024 2:13 PM
To: Chris Caffalette <ccaffalett@rockefeller.edu>; pett@cgl.ucsf.edu <pett@cgl.ucsf.edu>
Subject: Re: [ChimeraX] #14880: defattr Attribute File recipient value molecules or structures
Caution: External email
#14880: defattr Attribute File recipient value molecules or structures
----------------------------------------+----------------------
Reporter: ccaffalett@… | Owner: pett
Type: enhancement | Status: accepted
Priority: normal | Milestone:
Component: Structure Editing | Version:
Resolution: | Keywords:
Blocked By: | Blocking:
Notify when closed: | Platform: all
Project: ChimeraX |
----------------------------------------+----------------------
Changes (by pett):
* component: Unassigned => Structure Editing
* owner: (none) => pett
* platform: => all
* project: => ChimeraX
* status: new => accepted
* type: defect => enhancement
Comment:
Hi Chris,
The defattr file uses "molecules" to be backwards compatible with
files used with our previous Chimera program. With ChimeraX we prefer the
nomenclature "structures", which is why you see that in the Render By
Attribute interface. It would seem good to allow that same term in
defattr files (along with backwards-compatible "molecules").
Though the setattr command allows setting chain attributes, the
defattr file doesn't currently support it. It probably should. We don't
currently show chain attributes in the Render By Attribute tool basically
because there are no interesting chain attributes to render by. If there
was some compelling use case, that could be changed.
--Eric
Eric Pettersen
UCSF Computer Graphics Lab
--
Ticket URL: <https://urldefense.proofpoint.com/v2/url?u=https-3A__www.rbvi.ucsf.edu_trac_ChimeraX_ticket_14880-23comment-3A2&d=DwIGaQ&c=JeTkUgVztGMmhKYjxsy2rfoWYibK1YmxXez1G3oNStg&r=nn6NjO3O7Q3PXmOR60aUp-5a_7DeTCpr_5i98LXGSUA&m=p-uNjNyOY9bg-C0wDHT2tYUEKBHdh8zuwwcIZJnolHWigvh76jDYGeajjQxZBDSy&s=amwTnE9WpyP_2KTMfXctewrAZHyBwrkwtdFF18Q348s&e= >
ChimeraX <https://urldefense.proofpoint.com/v2/url?u=https-3A__www.rbvi.ucsf.edu_chimerax_&d=DwIGaQ&c=JeTkUgVztGMmhKYjxsy2rfoWYibK1YmxXez1G3oNStg&r=nn6NjO3O7Q3PXmOR60aUp-5a_7DeTCpr_5i98LXGSUA&m=p-uNjNyOY9bg-C0wDHT2tYUEKBHdh8zuwwcIZJnolHWigvh76jDYGeajjQxZBDSy&s=oQI6H2eSAyxSCmoc3yeglGWtI-uWD4kLmj8GCuFPH9Q&e= >
ChimeraX Issue Tracker
comment:4 by , 2 years ago
| Cc: | added |
|---|---|
| Resolution: | → fixed |
| Status: | accepted → closed |
Actually, the *code* already supports all the features I outlined, as well as allowing setting attributes for bonds and pseudobonds! It's just that the description in the help page needs updating.