#14829 closed defect (not a bug)
Problem opening pdb file
| Reported by: | Owned by: | Eric Pettersen | |
|---|---|---|---|
| Priority: | normal | Milestone: | |
| Component: | Input/Output | Version: | |
| Keywords: | Cc: | ||
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
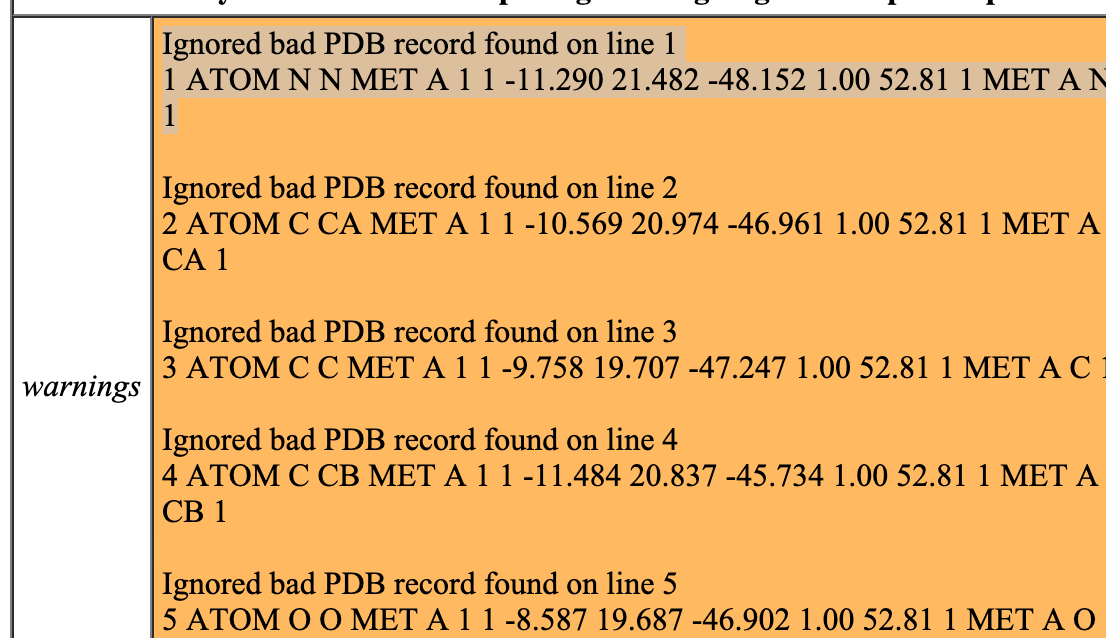
Hi, I have problems opening the attached file. I get the following warning messages (see screenshot) but unfortunately I cannot immediately see what’s wrong and therefore unable to find a fix. The first 5 lines of my pdb file are: 1 ATOM N N MET A 1 1 -11.290 21.482 -48.152 1.00 52.81 1 MET A N 1 2 ATOM C CA MET A 1 1 -10.569 20.974 -46.961 1.00 52.81 1 MET A CA 1 3 ATOM C C MET A 1 1 -9.758 19.707 -47.247 1.00 52.81 1 MET A C 1 4 ATOM C CB MET A 1 1 -11.484 20.837 -45.734 1.00 52.81 1 MET A CB 1 5 ATOM O O MET A 1 1 -8.587 19.687 -46.902 1.00 52.81 1 MET A O 1 I’d be grateful for your help George
Attachments (2)
Change History (7)
by , 2 years ago
| Attachment: | Screenshot 2024-03-26 at 23.25.30.png added |
|---|
comment:1 by , 2 years ago
| Component: | Unassigned → Input/Output |
|---|---|
| Owner: | set to |
| Platform: | → all |
| Project: | → ChimeraX |
| Status: | new → accepted |
comment:2 by , 2 years ago
| Resolution: | → not a bug |
|---|---|
| Status: | accepted → closed |
Hi George,
The first five columns of each line in your file are a number followed by whitespace -- then the remainder of the line looks like a legal PDB record. You need to get rid of those first 5 columns.
--Eric
Eric Pettersen
UCSF Computer Graphics Lab
comment:3 by , 2 years ago
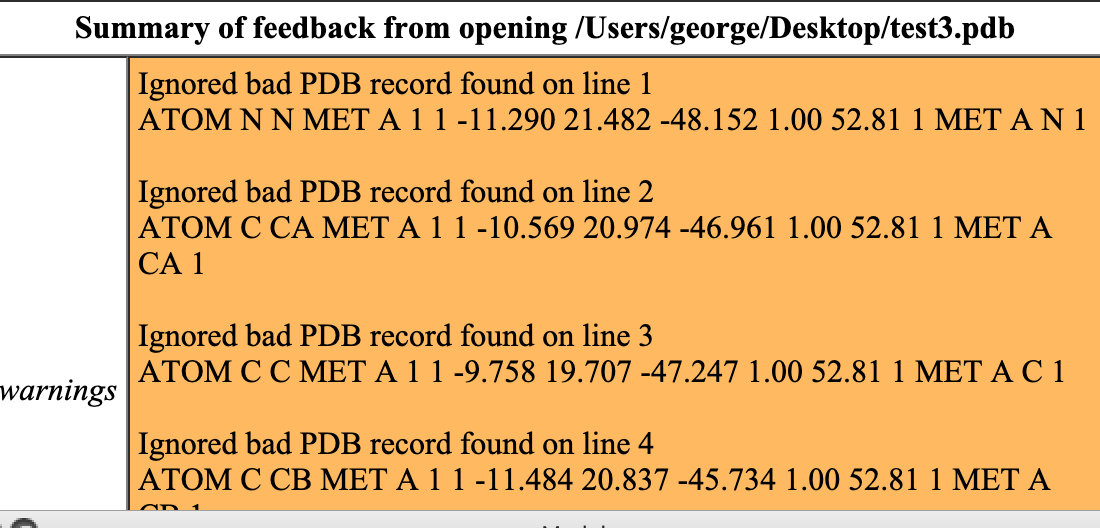
Eric, many thanks for the prompt reply. I got rid of the first five columns. I still get errors. My pdb file now looks like: ATOM N N MET A 1 1 -11.290 21.482 -48.152 1.00 52.81 1 MET A N 1 ATOM C CA MET A 1 1 -10.569 20.974 -46.961 1.00 52.81 1 MET A CA 1 ATOM C C MET A 1 1 -9.758 19.707 -47.247 1.00 52.81 1 MET A C 1 ATOM C CB MET A 1 1 -11.484 20.837 -45.734 1.00 52.81 1 MET A CB 1 ATOM O O MET A 1 1 -8.587 19.687 -46.902 1.00 52.81 1 MET A O 1 ATOM C CG MET A 1 1 -10.981 21.771 -44.629 1.00 52.81 1 MET A CG 1 ATOM S SD MET A 1 1 -11.969 21.700 -43.119 1.00 52.81 1 MET A SD 1 ATOM C CE MET A 1 1 -10.949 22.729 -42.026 1.00 52.81 1 MET A CE 1 ATOM N N LEU A 1 2 -10.309 18.685 -47.918 1.00 54.72 2 LEU A N 1 ATOM C CA LEU A 1 2 -9.550 17.472 -48.287 1.00 54.72 2 LEU A CA 1 And the error warnings are in the attached snapshot
Apologies for this George
comment:4 by , 2 years ago
Hi George, The PDB is a column-oriented format, so column position matters. The definition/format of an ATOM record is here: wwPDB Format version 3.3: Coordinate Section <https://www.wwpdb.org/documentation/file-format-content/format33/sect9.html#ATOM>. I guess the first five columns you had before were supposed to be the serial numbers. As per the ATOM definition, the serial number comes after "ATOM ". You can see the difference between your ATOM record (first line) and an ATOM record from 1gcn (second line): ATOM N N MET A 1 1 -11.290 21.482 -48.152 1.00 52.81 1 MET A N 1 ATOM 1 N HIS A 1 49.668 24.248 10.436 1.00 25.00 N The columns in your file need to line up with the columns in specified in the PDB format. --Eric
Note:
See TracTickets
for help on using tickets.
Added by email2trac