Opened 2 years ago
Last modified 2 years ago
#14598 assigned defect
Clipper session restore: self.cell and/or self.grid is None
| Reported by: | Owned by: | Tristan Croll | |
|---|---|---|---|
| Priority: | normal | Milestone: | |
| Component: | Third Party | Version: | |
| Keywords: | Cc: | Tom Goddard | |
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
The following bug report has been submitted:
Platform: Windows-10-10.0.19045
ChimeraX Version: 1.4 (2022-06-03 23:39:42 UTC)
Description
Trying to restore a session results in this error code being thrown
Log:
> preset "overall look" "publication 2 (depth-cued)"
Using preset: Overall Look / Publication 2 (Depth-Cued)
Preset expands to these ChimeraX commands:
set bg white
graphics silhouettes f
lighting depthCue t
> graphics silhouettes true
> lighting shadows false
> lighting depthCueStart 0.5
> lighting depthCueEnd 1.5
> lighting depthCueColor black
> graphics silhouettes width 1.5
UCSF ChimeraX version: 1.4 (2022-06-03)
© 2016-2022 Regents of the University of California. All rights reserved.
How to cite UCSF ChimeraX
> open "D:\OneDrive\Documents\University\University of Leicester PhD\23-24
> GAPDHA Paper\23-24 GAPDHA Paper\2023_GADPH_Final\GapA_NADP\GapA_NADP.cxs"
> format session
Unable to restore session, resetting.
Traceback (most recent call last):
File "D:\Programs\ChimeraX 1.4\bin\lib\site-
packages\chimerax\core\session.py", line 731, in restore
obj = sm.restore_snapshot(self, data)
File "C:\Users\Samuel\AppData\Local\UCSF\ChimeraX\1.4\site-
packages\chimerax\clipper\maps\xmapset.py", line 1019, in restore_snapshot
xmh = XmapHandler_Live(mapset, data['map name'],
File "C:\Users\Samuel\AppData\Local\UCSF\ChimeraX\1.4\site-
packages\chimerax\clipper\maps\xmapset.py", line 966, in __init__
super().__init__(mapset, name, is_difference_map=is_difference_map,
File "C:\Users\Samuel\AppData\Local\UCSF\ChimeraX\1.4\site-
packages\chimerax\clipper\maps\map_handler_base.py", line 256, in __init__
darray = self._generate_and_fill_data_array(bp.origin_xyz, bp.origin_grid,
bp.dim)
File "C:\Users\Samuel\AppData\Local\UCSF\ChimeraX\1.4\site-
packages\chimerax\clipper\maps\map_handler_base.py", line 384, in
_generate_and_fill_data_array
darray = self._generate_data_array(origin, grid_origin, dim)
File "C:\Users\Samuel\AppData\Local\UCSF\ChimeraX\1.4\site-
packages\chimerax\clipper\maps\map_handler_base.py", line 369, in
_generate_data_array
step = self.voxel_size, cell_angles = self.cell.angles_deg)
File "C:\Users\Samuel\AppData\Local\UCSF\ChimeraX\1.4\site-
packages\chimerax\clipper\maps\map_handler_base.py", line 306, in voxel_size
return self.cell.dim / self.grid.dim
AttributeError: 'NoneType' object has no attribute 'dim'
Exception ignored in: <function Drawing.__del__ at 0x000002A2C3172CA0>
Traceback (most recent call last):
File "D:\Programs\ChimeraX 1.4\bin\lib\site-
packages\chimerax\graphics\drawing.py", line 1187, in __del__
if not self.was_deleted:
AttributeError: 'XmapHandler_Live' object has no attribute 'was_deleted'
opened ChimeraX session
Traceback (most recent call last):
File "D:\Programs\ChimeraX 1.4\bin\lib\site-
packages\chimerax\core\triggerset.py", line 134, in invoke
return self._func(self._name, data)
File "C:\Users\Samuel\AppData\Local\UCSF\ChimeraX\1.4\site-
packages\chimerax\clipper\maps\xmapset.py", line 698, in
_recalculate_maps_if_needed
if self._recalc_needed and not xm.thread_running:
AttributeError: 'NoneType' object has no attribute 'thread_running'
Error processing trigger "new frame":
AttributeError: 'NoneType' object has no attribute 'thread_running'
File "C:\Users\Samuel\AppData\Local\UCSF\ChimeraX\1.4\site-
packages\chimerax\clipper\maps\xmapset.py", line 698, in
_recalculate_maps_if_needed
if self._recalc_needed and not xm.thread_running:
See log for complete Python traceback.
OpenGL version: 3.3.0 NVIDIA 536.23
OpenGL renderer: NVIDIA GeForce GTX 1070/PCIe/SSE2
OpenGL vendor: NVIDIA Corporation
Python: 3.9.11
Locale: en_GB.cp1252
Qt version: PyQt6 6.3.0, Qt 6.3.0
Qt runtime version: 6.3.0
Qt platform: windows
Manufacturer: System manufacturer
Model: System Product Name
OS: Microsoft Windows 10 Home (Build 19045)
Memory: 17,100,877,824
MaxProcessMemory: 137,438,953,344
CPU: 6 Intel(R) Core(TM) i5-8600K CPU @ 3.60GHz
OSLanguage: en-GB
Installed Packages:
alabaster: 0.7.12
appdirs: 1.4.4
Babel: 2.10.1
backcall: 0.2.0
blockdiag: 3.0.0
certifi: 2022.5.18.1
cftime: 1.6.0
charset-normalizer: 2.0.12
ChimeraX-AddCharge: 1.2.3
ChimeraX-AddH: 2.1.11
ChimeraX-AlignmentAlgorithms: 2.0
ChimeraX-AlignmentHdrs: 3.2.1
ChimeraX-AlignmentMatrices: 2.0
ChimeraX-Alignments: 2.4.3
ChimeraX-AlphaFold: 1.0
ChimeraX-AltlocExplorer: 1.0.2
ChimeraX-AmberInfo: 1.0
ChimeraX-Arrays: 1.0
ChimeraX-Atomic: 1.39.1
ChimeraX-AtomicLibrary: 7.0
ChimeraX-AtomSearch: 2.0.1
ChimeraX-AxesPlanes: 2.1
ChimeraX-BasicActions: 1.1
ChimeraX-BILD: 1.0
ChimeraX-BlastProtein: 2.1.1
ChimeraX-BondRot: 2.0
ChimeraX-BugReporter: 1.0
ChimeraX-BuildStructure: 2.7
ChimeraX-Bumps: 1.0
ChimeraX-BundleBuilder: 1.1
ChimeraX-ButtonPanel: 1.0
ChimeraX-CageBuilder: 1.0
ChimeraX-CellPack: 1.0
ChimeraX-Centroids: 1.2
ChimeraX-ChemGroup: 2.0
ChimeraX-Clashes: 2.2.4
ChimeraX-Clipper: 0.18.0
ChimeraX-ColorActions: 1.0
ChimeraX-ColorGlobe: 1.0
ChimeraX-ColorKey: 1.5.1
ChimeraX-CommandLine: 1.2.3
ChimeraX-ConnectStructure: 2.0.1
ChimeraX-Contacts: 1.0
ChimeraX-Core: 1.4
ChimeraX-CoreFormats: 1.1
ChimeraX-coulombic: 1.3.2
ChimeraX-Crosslinks: 1.0
ChimeraX-Crystal: 1.0
ChimeraX-CrystalContacts: 1.0
ChimeraX-DataFormats: 1.2.2
ChimeraX-Dicom: 1.1
ChimeraX-DistMonitor: 1.1.5
ChimeraX-Dssp: 2.0
ChimeraX-EMDB-SFF: 1.0
ChimeraX-ExperimentalCommands: 1.0
ChimeraX-FileHistory: 1.0
ChimeraX-FunctionKey: 1.0
ChimeraX-Geometry: 1.2
ChimeraX-gltf: 1.0
ChimeraX-Graphics: 1.1
ChimeraX-Hbonds: 2.1.2
ChimeraX-Help: 1.2
ChimeraX-HKCage: 1.3
ChimeraX-IHM: 1.1
ChimeraX-ImageFormats: 1.2
ChimeraX-IMOD: 1.0
ChimeraX-IO: 1.0.1
ChimeraX-ItemsInspection: 1.0
ChimeraX-Label: 1.1.1
ChimeraX-ListInfo: 1.1.1
ChimeraX-Log: 1.1.5
ChimeraX-LookingGlass: 1.1
ChimeraX-Maestro: 1.8.1
ChimeraX-Map: 1.1
ChimeraX-MapData: 2.0
ChimeraX-MapEraser: 1.0
ChimeraX-MapFilter: 2.0
ChimeraX-MapFit: 2.0
ChimeraX-MapSeries: 2.1
ChimeraX-Markers: 1.0
ChimeraX-Mask: 1.0
ChimeraX-MatchMaker: 2.0.6
ChimeraX-MDcrds: 2.6
ChimeraX-MedicalToolbar: 1.0.1
ChimeraX-Meeting: 1.0
ChimeraX-MLP: 1.1
ChimeraX-mmCIF: 2.7
ChimeraX-MMTF: 2.1
ChimeraX-Modeller: 1.5.5
ChimeraX-ModelPanel: 1.3.2
ChimeraX-ModelSeries: 1.0
ChimeraX-Mol2: 2.0
ChimeraX-Morph: 1.0
ChimeraX-MouseModes: 1.1
ChimeraX-Movie: 1.0
ChimeraX-Neuron: 1.0
ChimeraX-Nucleotides: 2.0.2
ChimeraX-OpenCommand: 1.9
ChimeraX-PDB: 2.6.6
ChimeraX-PDBBio: 1.0
ChimeraX-PDBLibrary: 1.0.2
ChimeraX-PDBMatrices: 1.0
ChimeraX-PickBlobs: 1.0
ChimeraX-Positions: 1.0
ChimeraX-PresetMgr: 1.1
ChimeraX-PubChem: 2.1
ChimeraX-ReadPbonds: 1.0.1
ChimeraX-Registration: 1.1
ChimeraX-RemoteControl: 1.0
ChimeraX-ResidueFit: 1.0
ChimeraX-RestServer: 1.1
ChimeraX-RNALayout: 1.0
ChimeraX-RotamerLibMgr: 2.0.1
ChimeraX-RotamerLibsDunbrack: 2.0
ChimeraX-RotamerLibsDynameomics: 2.0
ChimeraX-RotamerLibsRichardson: 2.0
ChimeraX-SaveCommand: 1.5.1
ChimeraX-SchemeMgr: 1.0
ChimeraX-SDF: 2.0
ChimeraX-Segger: 1.0
ChimeraX-Segment: 1.0
ChimeraX-SelInspector: 1.0
ChimeraX-SeqView: 2.6
ChimeraX-Shape: 1.0.1
ChimeraX-Shell: 1.0
ChimeraX-Shortcuts: 1.1
ChimeraX-ShowAttr: 1.0
ChimeraX-ShowSequences: 1.0
ChimeraX-SideView: 1.0
ChimeraX-Smiles: 2.1
ChimeraX-SmoothLines: 1.0
ChimeraX-SpaceNavigator: 1.0
ChimeraX-StdCommands: 1.8
ChimeraX-STL: 1.0
ChimeraX-Storm: 1.0
ChimeraX-StructMeasure: 1.0.1
ChimeraX-Struts: 1.0.1
ChimeraX-Surface: 1.0
ChimeraX-SwapAA: 2.0
ChimeraX-SwapRes: 2.1.1
ChimeraX-TapeMeasure: 1.0
ChimeraX-Test: 1.0
ChimeraX-Toolbar: 1.1.1
ChimeraX-ToolshedUtils: 1.2.1
ChimeraX-Tug: 1.0
ChimeraX-UI: 1.18.3
ChimeraX-uniprot: 2.2
ChimeraX-UnitCell: 1.0
ChimeraX-ViewDockX: 1.1.2
ChimeraX-VIPERdb: 1.0
ChimeraX-Vive: 1.1
ChimeraX-VolumeMenu: 1.0
ChimeraX-VTK: 1.0
ChimeraX-WavefrontOBJ: 1.0
ChimeraX-WebCam: 1.0
ChimeraX-WebServices: 1.1.0
ChimeraX-Zone: 1.0
colorama: 0.4.4
comtypes: 1.1.10
cxservices: 1.2
cycler: 0.11.0
Cython: 0.29.26
debugpy: 1.6.0
decorator: 5.1.1
docutils: 0.17.1
entrypoints: 0.4
filelock: 3.4.2
fonttools: 4.33.3
funcparserlib: 1.0.0
grako: 3.16.5
h5py: 3.7.0
html2text: 2020.1.16
idna: 3.3
ihm: 0.27
imagecodecs: 2021.11.20
imagesize: 1.3.0
ipykernel: 6.6.1
ipython: 7.31.1
ipython-genutils: 0.2.0
jedi: 0.18.1
Jinja2: 3.0.3
jupyter-client: 7.1.0
jupyter-core: 4.10.0
kiwisolver: 1.4.2
line-profiler: 3.4.0
lxml: 4.7.1
lz4: 3.1.10
MarkupSafe: 2.1.1
matplotlib: 3.5.1
matplotlib-inline: 0.1.3
msgpack: 1.0.3
nest-asyncio: 1.5.5
netCDF4: 1.5.8
networkx: 2.6.3
numexpr: 2.8.1
numpy: 1.22.1
openvr: 1.16.802
packaging: 21.3
ParmEd: 3.4.3
parso: 0.8.3
pickleshare: 0.7.5
Pillow: 9.0.1
pip: 21.3.1
pkginfo: 1.8.2
prompt-toolkit: 3.0.29
psutil: 5.9.0
pycollada: 0.7.2
pydicom: 2.2.2
Pygments: 2.11.2
PyOpenGL: 3.1.5
PyOpenGL-accelerate: 3.1.5
pyparsing: 3.0.9
PyQt6-commercial: 6.3.0
PyQt6-Qt6: 6.3.0
PyQt6-sip: 13.3.1
PyQt6-WebEngine-commercial: 6.3.0
PyQt6-WebEngine-Qt6: 6.3.0
python-dateutil: 2.8.2
pytz: 2022.1
pywin32: 303
pyzmq: 23.1.0
qtconsole: 5.3.0
QtPy: 2.1.0
RandomWords: 0.3.0
requests: 2.27.1
scipy: 1.7.3
setuptools: 59.8.0
sfftk-rw: 0.7.2
six: 1.16.0
snowballstemmer: 2.2.0
sortedcontainers: 2.4.0
Sphinx: 4.3.2
sphinx-autodoc-typehints: 1.15.2
sphinxcontrib-applehelp: 1.0.2
sphinxcontrib-blockdiag: 3.0.0
sphinxcontrib-devhelp: 1.0.2
sphinxcontrib-htmlhelp: 2.0.0
sphinxcontrib-jsmath: 1.0.1
sphinxcontrib-qthelp: 1.0.3
sphinxcontrib-serializinghtml: 1.1.5
suds-community: 1.0.0
tables: 3.7.0
tifffile: 2021.11.2
tinyarray: 1.2.4
tornado: 6.1
traitlets: 5.1.1
urllib3: 1.26.9
wcwidth: 0.2.5
webcolors: 1.11.1
wheel: 0.37.1
wheel-filename: 1.3.0
WMI: 1.5.1
File attachment: GapA_NADP.cxs
Attachments (2)
Change History (10)
by , 2 years ago
| Attachment: | GapA_NADP.cxs added |
|---|
comment:1 by , 2 years ago
| Cc: | added |
|---|---|
| Component: | Unassigned → Third Party |
| Owner: | set to |
| Platform: | → all |
| Project: | → ChimeraX |
| Status: | new → assigned |
| Summary: | ChimeraX bug report submission → Clipper session restore: self.cell and/or self.grid is None |
Reported by Samuel
comment:2 by , 2 years ago
That's a rather old version of ChimeraX and ISOLDE... I just tried opening the session file in the current ISOLDE build and got the same error. Could you (a) share a little more detail about what was in the session (i.e. what type(s) of map(s)) and (b) try recreating it in the latest version and see if you get the same error after saving and re-opening the session?
comment:3 by , 2 years ago
Hi there,
yes absolutely, the maps are directly from refmac5 - I believe when I saved it I had closed all but the live 2fo-fc map (can't remember the exact name but that is what it represents)
I can now open sessions in the updated version but not one with maps - this is the error message I get,
Unable to restore session, resetting.
Traceback (most recent call last):
File "D:\Programs\ChimeraX\bin\Lib\site-packages\chimerax\core\session.py", line 747, in restore
obj = sm.restore_snapshot(self, data)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "C:\Users\Samuel\AppData\Local\UCSF\ChimeraX\1.7\Python311\site-packages\chimerax\clipper\maps\xmapset.py", line 1039, in restore_snapshot
xmh = XmapHandler_Live(mapset, data['map name'],
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "C:\Users\Samuel\AppData\Local\UCSF\ChimeraX\1.7\Python311\site-packages\chimerax\clipper\maps\xmapset.py", line 986, in __init__
super().__init__(mapset, name, is_difference_map=is_difference_map,
File "C:\Users\Samuel\AppData\Local\UCSF\ChimeraX\1.7\Python311\site-packages\chimerax\clipper\maps\map_handler_base.py", line 256, in __init__
darray = self._generate_and_fill_data_array(bp.origin_xyz, bp.origin_grid, bp.dim)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "C:\Users\Samuel\AppData\Local\UCSF\ChimeraX\1.7\Python311\site-packages\chimerax\clipper\maps\map_handler_base.py", line 384, in _generate_and_fill_data_array
darray = self._generate_data_array(origin, grid_origin, dim)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "C:\Users\Samuel\AppData\Local\UCSF\ChimeraX\1.7\Python311\site-packages\chimerax\clipper\maps\map_handler_base.py", line 369, in _generate_data_array
step = self.voxel_size, cell_angles = self.cell.angles_deg)
^^^^^^^^^^^^^^^
File "C:\Users\Samuel\AppData\Local\UCSF\ChimeraX\1.7\Python311\site-packages\chimerax\clipper\maps\map_handler_base.py", line 306, in voxel_size
return self.cell.dim / self.grid.dim
^^^^^^^^^^^^^
AttributeError: 'NoneType' object has no attribute 'dim'
UCSF ChimeraX version: 1.7.1 (2024-01-23)
bw,
Samuel Foster MBioChem/ Molecular and Cell Biology Research PhD Student
________________________________
From: ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu>
Sent: 16 February 2024 17:28
To: tcroll@altoslabs.com <tcroll@altoslabs.com>; Foster, Samuel <sf314@leicester.ac.uk>
Cc: goddard@cgl.ucsf.edu <goddard@cgl.ucsf.edu>
Subject: Re: [ChimeraX] #14598: Clipper session restore: self.cell and/or self.grid is None
[You don't often get email from chimerax-bugs-admin@cgl.ucsf.edu. Learn why this is important at https://aka.ms/LearnAboutSenderIdentification ]
***CAUTION:*** This email was sent from an EXTERNAL source. Think before clicking links or opening attachments.
#14598: Clipper session restore: self.cell and/or self.grid is None
----------------------------------+---------------------------
Reporter: sf314@… | Owner: Tristan Croll
Type: defect | Status: assigned
Priority: normal | Milestone:
Component: Third Party | Version:
Resolution: | Keywords:
Blocked By: | Blocking:
Notify when closed: | Platform: all
Project: ChimeraX |
----------------------------------+---------------------------
Comment (by Tristan Croll):
That's a rather old version of ChimeraX and ISOLDE... I just tried opening
the session file in the current ISOLDE build and got the same error. Could
you (a) share a little more detail about what was in the session (i.e.
what type(s) of map(s)) and (b) try recreating it in the latest version
and see if you get the same error after saving and re-opening the session?
--
Ticket URL: <https://eur03.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.rbvi.ucsf.edu%2Ftrac%2FChimeraX%2Fticket%2F14598%23comment%3A2&data=05%7C02%7Csf314%40leicester.ac.uk%7C9b9a69ac563d4b72e0ca08dc2f14c5de%7Caebecd6a31d44b0195ce8274afe853d9%7C0%7C0%7C638437013533794020%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C0%7C%7C%7C&sdata=mRdWG1j0lEzmdWVoIl5PufHErXICZKHPG7WI9j2Xwss%3D&reserved=0<https://www.rbvi.ucsf.edu/trac/ChimeraX/ticket/14598#comment:2>>
ChimeraX <https://eur03.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.rbvi.ucsf.edu%2Fchimerax%2F&data=05%7C02%7Csf314%40leicester.ac.uk%7C9b9a69ac563d4b72e0ca08dc2f14c5de%7Caebecd6a31d44b0195ce8274afe853d9%7C0%7C0%7C638437013533801363%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C0%7C%7C%7C&sdata=Ed19uL1wdKQPpbaELqIQzPVk6hZPaFrf4U7G%2BSKPL60%3D&reserved=0<https://www.rbvi.ucsf.edu/chimerax/>>
ChimeraX Issue Tracker
comment:4 by , 2 years ago
Ah - that was the hint I needed to replicate the problem. Looks like you deleted the model called "Reflection Data". To be perfectly honest, in (many years after the fact) hindsight I should have never made that an actual Model object - at the time I had some half-baked plan to make the HKLs viewable in the main window, but that never really got off the ground. As the name suggests, that's hosting the experimental data that the maps are actually calculated from, so closing it kills all future map recalculations (I get the same error if I delete it then move an atom to trigger recalculation). So the bad news is your session file is going to be unrecoverable without a bit of effort (if you have work in there that will take a long time to replicate, let me know and I'll see what I can do). The good news is that to avoid it in future, all you have to do is not close the "Reflection Data" model.
I guess it's probably time I gave this part of the data layout a bit of a rethink, but I can't promise anything immediate - most of my working time is taken up by other duties these days.
comment:5 by , 2 years ago
Many thanks for your quick reply!
Okay that makes sense I hadn't realised that is what that reflection data model was doing - I shall do that in the future.
I was able to reproduce the figure as I had learnt all the commands needed (just about- I've only been using ChimeraX a week but downloaded it a while ago when I learnt of it to see the comparison with Chimera, hence the older version), so don't worry about trying to recover this particular session,
And yes I am sure development takes a long time, so i'm very grateful you were able to get back to me so soon!
bw,
Samuel Foster MBioChem/ Molecular and Cell Biology Research PhD Student
________________________________
From: ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu>
Sent: 19 February 2024 12:32
To: Foster, Samuel <sf314@leicester.ac.uk>; tcroll@altoslabs.com <tcroll@altoslabs.com>
Cc: goddard@cgl.ucsf.edu <goddard@cgl.ucsf.edu>
Subject: Re: [ChimeraX] #14598: Clipper session restore: self.cell and/or self.grid is None
[You don't often get email from chimerax-bugs-admin@cgl.ucsf.edu. Learn why this is important at https://aka.ms/LearnAboutSenderIdentification ]
***CAUTION:*** This email was sent from an EXTERNAL source. Think before clicking links or opening attachments.
#14598: Clipper session restore: self.cell and/or self.grid is None
----------------------------------+---------------------------
Reporter: sf314@… | Owner: Tristan Croll
Type: defect | Status: assigned
Priority: normal | Milestone:
Component: Third Party | Version:
Resolution: | Keywords:
Blocked By: | Blocking:
Notify when closed: | Platform: all
Project: ChimeraX |
----------------------------------+---------------------------
Comment (by Tristan Croll):
Ah - that was the hint I needed to replicate the problem. Looks like you
deleted the model called "Reflection Data". To be perfectly honest, in
(many years after the fact) hindsight I should have never made that an
actual Model object - at the time I had some half-baked plan to make the
HKLs viewable in the main window, but that never really got off the
ground. As the name suggests, that's hosting the experimental data that
the maps are actually calculated from, so closing it kills all future map
recalculations (I get the same error if I delete it then move an atom to
trigger recalculation). So the bad news is your session file is going to
be unrecoverable without a bit of effort (if you have work in there that
will take a long time to replicate, let me know and I'll see what I can
do). The good news is that to avoid it in future, all you have to do is
not close the "Reflection Data" model.
I guess it's probably time I gave this part of the data layout a bit of a
rethink, but I can't promise anything immediate - most of my working time
is taken up by other duties these days.
--
Ticket URL: <https://eur03.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.rbvi.ucsf.edu%2Ftrac%2FChimeraX%2Fticket%2F14598%23comment%3A4&data=05%7C02%7Csf314%40leicester.ac.uk%7Ca968fa69dc36420dd59308dc3146e10f%7Caebecd6a31d44b0195ce8274afe853d9%7C0%7C0%7C638439427702870733%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C0%7C%7C%7C&sdata=bU6AeTyl1kbRknrtoNGAZQ9JlhFDmYNIzGFVOnn9Pkw%3D&reserved=0<https://www.rbvi.ucsf.edu/trac/ChimeraX/ticket/14598#comment:4>>
ChimeraX <https://eur03.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.rbvi.ucsf.edu%2Fchimerax%2F&data=05%7C02%7Csf314%40leicester.ac.uk%7Ca968fa69dc36420dd59308dc3146e10f%7Caebecd6a31d44b0195ce8274afe853d9%7C0%7C0%7C638439427702880082%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C0%7C%7C%7C&sdata=MrQvCL%2F4MCh15jKHTevpY%2FA7fhTiQ0mN3LA5hUMpWf8%3D&reserved=0<https://www.rbvi.ucsf.edu/chimerax/>>
ChimeraX Issue Tracker
comment:6 by , 2 years ago
OK - glad to know there wasn't anything critical in there! On Mon, Feb 19, 2024 at 12:39 PM ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu> wrote: > > > > > > > > > > > >
comment:7 by , 2 years ago
Sorry this is a little off topic,
Is there an equivalent command to ~set singleLayer<https://www.cgl.ucsf.edu/chimera/docs/UsersGuide/midas/set.html#singlelayer> from Chimera? I am trying to show the 2Fo-Fc map and Anaomlous difference map at the same time but the 2Fo-Fc is displayed over the top when both are set to transparent surfaces.
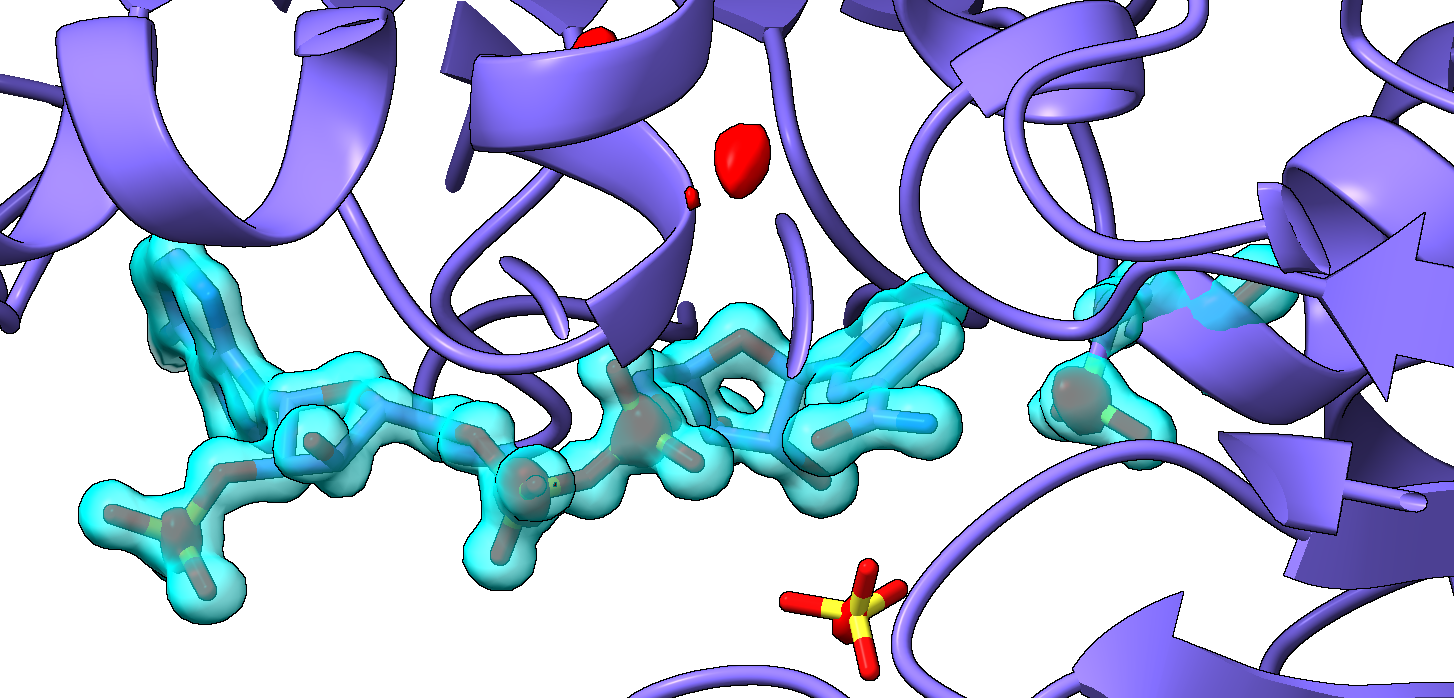
[cid:308f612b-3f38-4aa3-bcac-2e8596c4a812]
blue is the 2Fo-Fc, Red is anomalous signal. I would like to see both and the atoms underneath akin to this mockup
[Image]
Many thanks,
Samuel Foster MBioChem/ Molecular and Cell Biology Research PhD Student
________________________________
From: ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu>
Sent: 19 February 2024 14:22
To: tcroll@altoslabs.com <tcroll@altoslabs.com>; Foster, Samuel <sf314@leicester.ac.uk>
Cc: goddard@cgl.ucsf.edu <goddard@cgl.ucsf.edu>
Subject: Re: [ChimeraX] #14598: Clipper session restore: self.cell and/or self.grid is None
[You don't often get email from chimerax-bugs-admin@cgl.ucsf.edu. Learn why this is important at https://aka.ms/LearnAboutSenderIdentification ]
***CAUTION:*** This email was sent from an EXTERNAL source. Think before clicking links or opening attachments.
#14598: Clipper session restore: self.cell and/or self.grid is None
----------------------------------+---------------------------
Reporter: sf314@… | Owner: Tristan Croll
Type: defect | Status: assigned
Priority: normal | Milestone:
Component: Third Party | Version:
Resolution: | Keywords:
Blocked By: | Blocking:
Notify when closed: | Platform: all
Project: ChimeraX |
----------------------------------+---------------------------
Comment (by Tristan Croll):
{{{
OK - glad to know there wasn't anything critical in there!
On Mon, Feb 19, 2024 at 12:39 PM ChimeraX <ChimeraX-bugs-
admin@cgl.ucsf.edu>
wrote:
>
>
>
>
>
>
>
>
>
>
>
>
}}}
--
Ticket URL: <https://eur03.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.rbvi.ucsf.edu%2Ftrac%2FChimeraX%2Fticket%2F14598%23comment%3A6&data=05%7C02%7Csf314%40leicester.ac.uk%7C706ea4cc8f8a4c5fd80408dc31563e1f%7Caebecd6a31d44b0195ce8274afe853d9%7C0%7C0%7C638439493747145280%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C0%7C%7C%7C&sdata=yCAyXObKOpd8XKTh3O9G3N9Wr5jJYU8yS%2BHsYdt5fzM%3D&reserved=0<https://www.rbvi.ucsf.edu/trac/ChimeraX/ticket/14598#comment:6>>
ChimeraX <https://eur03.safelinks.protection.outlook.com/?url=https%3A%2F%2Fwww.rbvi.ucsf.edu%2Fchimerax%2F&data=05%7C02%7Csf314%40leicester.ac.uk%7C706ea4cc8f8a4c5fd80408dc31563e1f%7Caebecd6a31d44b0195ce8274afe853d9%7C0%7C0%7C638439493747152742%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C0%7C%7C%7C&sdata=QpmSuAAoxxHbPn%2BISS7zfNgeuI65iZObZoOpt1Hs2EU%3D&reserved=0<https://www.rbvi.ucsf.edu/chimerax/>>
ChimeraX Issue Tracker
comment:8 by , 2 years ago
At the moment I’m afraid it’s impossible to overlay two transparent surfaces in ChimeraX - only the foreground one will show. To get the effect you want, you’ll need to use wireframes for either all or all-but-one of the maps. On Tue, 20 Feb 2024 at 12:24, ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu> wrote: > > >
Added by email2trac