Opened 2 years ago
Closed 2 years ago
#14554 closed defect (limitation)
Small checkboxes on second monitor
| Reported by: | Owned by: | Eric Pettersen | |
|---|---|---|---|
| Priority: | normal | Milestone: | |
| Component: | UI | Version: | |
| Keywords: | Cc: | Tom Goddard | |
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
The following bug report has been submitted:
Platform: Windows-10-10.0.22631
ChimeraX Version: 1.7.1 (2024-01-23 01:58:08 UTC)
Description
Hi,
When opening Chimera, the boxes in the model browser to select and show models are fine, but when moving the window to a new screen with a different resolution, they become tiny. Can this be fixed somehow?
Many thanks
Jakob Andersson
Log:
Startup Messages
---
warning | Replacing fetcher for 'pdb_nmr' and format NMRSTAR from NMRSTAR
bundle with that from NMRSTAR bundle
UCSF ChimeraX version: 1.7.1 (2024-01-23)
© 2016-2023 Regents of the University of California. All rights reserved.
How to cite UCSF ChimeraX
> open "C:/Users/mrand/OneDrive/Work/ISTA/Data/Yarrowia CI/Model
> building/renamed maps/composite maps/open 1/T128_open1_PA_j688.mrc"
Opened T128_open1_PA_j688.mrc as #1, grid size 588,588,588, pixel 1.06, shown
at level 0.00355, step 4, values float32
> open "C:/Users/mrand/OneDrive/Work/ISTA/Data/Yarrowia CI/Model
> building/renamed maps/composite maps/open 1/T128_open1_MA_j685.mrc"
Opened T128_open1_MA_j685.mrc as #2, grid size 588,588,588, pixel 1.06, shown
at level 0.00343, step 4, values float32
> open "C:/Users/mrand/OneDrive/Work/ISTA/Data/Yarrowia CI/Model
> building/renamed maps/composite maps/open 1/T128_open1_global_j669.mrc"
Opened T128_open1_global_j669.mrc as #3, grid size 588,588,588, pixel 1.06,
shown at level 0.00319, step 4, values float32
> volume #1-5 step 1
> volume #1-5 level 0.015
> hide #!1 models
> hide #!2 models
> hide #!3 models
> open "C:/Users/mrand/OneDrive/Work/ISTA/Data/Yarrowia CI/Model
> building/Structure comparison chimera sessions/MA-only_glacios-
> model_phenix_run3.pdb"
Chain information for MA-only_glacios-model_phenix_run3.pdb #4
---
Chain | Description
A | No description available
D | No description available
H | No description available
I | No description available
J | No description available
K | No description available
L | No description available
M | No description available
N | No description available
O | No description available
U | No description available
X | No description available
Y | No description available
Z | No description available
a | No description available
b | No description available
d | No description available
e | No description available
g | No description available
h | No description available
i | No description available
j | No description available
k | No description available
l | No description available
m | No description available
n | No description available
o | No description available
p | No description available
> open "C:/Users/mrand/OneDrive/Work/ISTA/Data/Yarrowia CI/Model
> building/Structure comparison chimera sessions/PA-only_open2_glacios-
> model_phenix_run3.pdb"
Chain information for PA-only_open2_glacios-model_phenix_run3.pdb #5
---
Chain | Description
A | No description available
B | No description available
C | No description available
D | No description available
E | No description available
F | No description available
G | No description available
H | No description available
I | No description available
J | No description available
P | No description available
Q | No description available
R | No description available
S | No description available
T | No description available
V | No description available
W | No description available
Z | No description available
a | No description available
b | No description available
c | No description available
f | No description available
> hide #!4 models
> hide #!5 models
> show #!4 models
> hide #!4 models
> show #!4 models
> show #!5 models
> hide #!5 models
> show #!5 models
> hide #!5 models
> show #!5 models
> select add #4
34390 atoms, 35036 bonds, 10 pseudobonds, 4336 residues, 2 models selected
> select add #5
65497 atoms, 66861 bonds, 18 pseudobonds, 8285 residues, 4 models selected
> hide (#!4-5 & sel) target a
> cartoon (#!4-5 & sel)
> ui mousemode right select
> select clear
> select add #5
31107 atoms, 31825 bonds, 8 pseudobonds, 3949 residues, 2 models selected
> select subtract #5
Nothing selected
> select add #5
31107 atoms, 31825 bonds, 8 pseudobonds, 3949 residues, 2 models selected
> select subtract #5
Nothing selected
> hide #5.1 models
> hide #!5 models
> show #!5 models
> show #5.1 models
> hide #5.1 models
> show #!1 models
> hide #!1 models
> view #1 clip false
No displayed objects specified.
> set bgColor black
> set bgColor transparent
OpenGL version: 3.3.0 - Build 31.0.101.4502
OpenGL renderer: Intel(R) Iris(R) Xe Graphics
OpenGL vendor: Intel
Python: 3.11.2
Locale: en_AT.cp1252
Qt version: PyQt6 6.3.1, Qt 6.3.1
Qt runtime version: 6.3.2
Qt platform: windows
Manufacturer: Microsoft Corporation
Model: Surface Pro 9
OS: Microsoft Windows 11 Home (Build 22631)
Memory: 8,405,700,608
MaxProcessMemory: 137,438,953,344
CPU: 12 12th Gen Intel(R) Core(TM) i5-1235U
OSLanguage: en-GB
Installed Packages:
alabaster: 0.7.16
appdirs: 1.4.4
asttokens: 2.4.1
Babel: 2.14.0
backcall: 0.2.0
beautifulsoup4: 4.11.2
blockdiag: 3.0.0
blosc2: 2.0.0
build: 0.10.0
certifi: 2023.11.17
cftime: 1.6.3
charset-normalizer: 3.3.2
ChimeraX-AddCharge: 1.5.13
ChimeraX-AddH: 2.2.5
ChimeraX-AlignmentAlgorithms: 2.0.1
ChimeraX-AlignmentHdrs: 3.4.1
ChimeraX-AlignmentMatrices: 2.1
ChimeraX-Alignments: 2.12.2
ChimeraX-AlphaFold: 1.0
ChimeraX-AltlocExplorer: 1.1.1
ChimeraX-AmberInfo: 1.0
ChimeraX-Arrays: 1.1
ChimeraX-Atomic: 1.49.1
ChimeraX-AtomicLibrary: 12.1.5
ChimeraX-AtomSearch: 2.0.1
ChimeraX-AxesPlanes: 2.3.2
ChimeraX-BasicActions: 1.1.2
ChimeraX-BILD: 1.0
ChimeraX-BlastProtein: 2.1.2
ChimeraX-BondRot: 2.0.4
ChimeraX-BugReporter: 1.0.1
ChimeraX-BuildStructure: 2.10.5
ChimeraX-Bumps: 1.0
ChimeraX-BundleBuilder: 1.2.2
ChimeraX-ButtonPanel: 1.0.1
ChimeraX-CageBuilder: 1.0.1
ChimeraX-CellPack: 1.0
ChimeraX-Centroids: 1.3.2
ChimeraX-ChangeChains: 1.1
ChimeraX-CheckWaters: 1.3.2
ChimeraX-ChemGroup: 2.0.1
ChimeraX-Clashes: 2.2.4
ChimeraX-ColorActions: 1.0.3
ChimeraX-ColorGlobe: 1.0
ChimeraX-ColorKey: 1.5.5
ChimeraX-CommandLine: 1.2.5
ChimeraX-ConnectStructure: 2.0.1
ChimeraX-Contacts: 1.0.1
ChimeraX-Core: 1.7.1
ChimeraX-CoreFormats: 1.2
ChimeraX-coulombic: 1.4.2
ChimeraX-Crosslinks: 1.0
ChimeraX-Crystal: 1.0
ChimeraX-CrystalContacts: 1.0.1
ChimeraX-DataFormats: 1.2.3
ChimeraX-Dicom: 1.2
ChimeraX-DistMonitor: 1.4
ChimeraX-DockPrep: 1.1.3
ChimeraX-Dssp: 2.0
ChimeraX-EMDB-SFF: 1.0
ChimeraX-ESMFold: 1.0
ChimeraX-FileHistory: 1.0.1
ChimeraX-FunctionKey: 1.0.1
ChimeraX-Geometry: 1.3
ChimeraX-gltf: 1.0
ChimeraX-Graphics: 1.1.1
ChimeraX-Hbonds: 2.4
ChimeraX-Help: 1.2.2
ChimeraX-HKCage: 1.3
ChimeraX-IHM: 1.1
ChimeraX-ImageFormats: 1.2
ChimeraX-IMOD: 1.0
ChimeraX-IO: 1.0.1
ChimeraX-ItemsInspection: 1.0.1
ChimeraX-IUPAC: 1.0
ChimeraX-Label: 1.1.8
ChimeraX-ListInfo: 1.2.2
ChimeraX-Log: 1.1.6
ChimeraX-LookingGlass: 1.1
ChimeraX-Maestro: 1.9.1
ChimeraX-Map: 1.1.4
ChimeraX-MapData: 2.0
ChimeraX-MapEraser: 1.0.1
ChimeraX-MapFilter: 2.0.1
ChimeraX-MapFit: 2.0
ChimeraX-MapSeries: 2.1.1
ChimeraX-Markers: 1.0.1
ChimeraX-Mask: 1.0.2
ChimeraX-MatchMaker: 2.1.2
ChimeraX-MCopy: 1.0
ChimeraX-MDcrds: 2.6.1
ChimeraX-MedicalToolbar: 1.0.2
ChimeraX-Meeting: 1.0.1
ChimeraX-MLP: 1.1.1
ChimeraX-mmCIF: 2.12.1
ChimeraX-MMTF: 2.2
ChimeraX-Modeller: 1.5.14
ChimeraX-ModelPanel: 1.4
ChimeraX-ModelSeries: 1.0.1
ChimeraX-Mol2: 2.0.3
ChimeraX-Mole: 1.0
ChimeraX-Morph: 1.0.2
ChimeraX-MouseModes: 1.2
ChimeraX-Movie: 1.0
ChimeraX-Neuron: 1.0
ChimeraX-Nifti: 1.1
ChimeraX-NRRD: 1.1
ChimeraX-Nucleotides: 2.0.3
ChimeraX-OpenCommand: 1.13.1
ChimeraX-PDB: 2.7.3
ChimeraX-PDBBio: 1.0.1
ChimeraX-PDBLibrary: 1.0.4
ChimeraX-PDBMatrices: 1.0
ChimeraX-PickBlobs: 1.0.1
ChimeraX-Positions: 1.0
ChimeraX-PresetMgr: 1.1
ChimeraX-PubChem: 2.1
ChimeraX-ReadPbonds: 1.0.1
ChimeraX-Registration: 1.1.2
ChimeraX-RemoteControl: 1.0
ChimeraX-RenderByAttr: 1.1
ChimeraX-RenumberResidues: 1.1
ChimeraX-ResidueFit: 1.0.1
ChimeraX-RestServer: 1.2
ChimeraX-RNALayout: 1.0
ChimeraX-RotamerLibMgr: 4.0
ChimeraX-RotamerLibsDunbrack: 2.0
ChimeraX-RotamerLibsDynameomics: 2.0
ChimeraX-RotamerLibsRichardson: 2.0
ChimeraX-SaveCommand: 1.5.1
ChimeraX-SchemeMgr: 1.0
ChimeraX-SDF: 2.0.2
ChimeraX-Segger: 1.0
ChimeraX-Segment: 1.0.1
ChimeraX-SelInspector: 1.0
ChimeraX-SeqView: 2.11
ChimeraX-Shape: 1.0.1
ChimeraX-Shell: 1.0.1
ChimeraX-Shortcuts: 1.1.1
ChimeraX-ShowSequences: 1.0.2
ChimeraX-SideView: 1.0.1
ChimeraX-Smiles: 2.1.2
ChimeraX-SmoothLines: 1.0
ChimeraX-SpaceNavigator: 1.0
ChimeraX-StdCommands: 1.12.4
ChimeraX-STL: 1.0.1
ChimeraX-Storm: 1.0
ChimeraX-StructMeasure: 1.1.2
ChimeraX-Struts: 1.0.1
ChimeraX-Surface: 1.0.1
ChimeraX-SwapAA: 2.0.1
ChimeraX-SwapRes: 2.2.2
ChimeraX-TapeMeasure: 1.0
ChimeraX-TaskManager: 1.0
ChimeraX-Test: 1.0
ChimeraX-Toolbar: 1.1.2
ChimeraX-ToolshedUtils: 1.2.4
ChimeraX-Topography: 1.0
ChimeraX-ToQuest: 1.0
ChimeraX-Tug: 1.0.1
ChimeraX-UI: 1.33.3
ChimeraX-uniprot: 2.3
ChimeraX-UnitCell: 1.0.1
ChimeraX-ViewDockX: 1.3.2
ChimeraX-VIPERdb: 1.0
ChimeraX-Vive: 1.1
ChimeraX-VolumeMenu: 1.0.1
ChimeraX-vrml: 1.0
ChimeraX-VTK: 1.0
ChimeraX-WavefrontOBJ: 1.0
ChimeraX-WebCam: 1.0.2
ChimeraX-WebServices: 1.1.3
ChimeraX-Zone: 1.0.1
colorama: 0.4.6
comm: 0.2.1
comtypes: 1.1.14
contourpy: 1.2.0
cxservices: 1.2.2
cycler: 0.12.1
Cython: 0.29.33
debugpy: 1.8.0
decorator: 5.1.1
docutils: 0.19
executing: 2.0.1
filelock: 3.9.0
fonttools: 4.47.2
funcparserlib: 2.0.0a0
glfw: 2.6.4
grako: 3.16.5
h5py: 3.10.0
html2text: 2020.1.16
idna: 3.6
ihm: 0.38
imagecodecs: 2023.9.18
imagesize: 1.4.1
ipykernel: 6.23.2
ipython: 8.14.0
ipython-genutils: 0.2.0
ipywidgets: 8.1.1
jedi: 0.18.2
Jinja2: 3.1.2
jupyter-client: 8.2.0
jupyter-core: 5.7.1
jupyterlab-widgets: 3.0.9
kiwisolver: 1.4.5
line-profiler: 4.0.2
lxml: 4.9.2
lz4: 4.3.2
MarkupSafe: 2.1.4
matplotlib: 3.7.2
matplotlib-inline: 0.1.6
msgpack: 1.0.4
nest-asyncio: 1.6.0
netCDF4: 1.6.2
networkx: 3.1
nibabel: 5.0.1
nptyping: 2.5.0
numexpr: 2.8.8
numpy: 1.25.1
openvr: 1.23.701
packaging: 23.2
ParmEd: 3.4.3
parso: 0.8.3
pep517: 0.13.0
pickleshare: 0.7.5
pillow: 10.2.0
pip: 23.0
pkginfo: 1.9.6
platformdirs: 4.1.0
prompt-toolkit: 3.0.43
psutil: 5.9.5
pure-eval: 0.2.2
py-cpuinfo: 9.0.0
pycollada: 0.7.2
pydicom: 2.3.0
Pygments: 2.16.1
pynrrd: 1.0.0
PyOpenGL: 3.1.7
PyOpenGL-accelerate: 3.1.7
pyopenxr: 1.0.2801
pyparsing: 3.0.9
pyproject-hooks: 1.0.0
PyQt6-commercial: 6.3.1
PyQt6-Qt6: 6.3.2
PyQt6-sip: 13.4.0
PyQt6-WebEngine-commercial: 6.3.1
PyQt6-WebEngine-Qt6: 6.3.2
python-dateutil: 2.8.2
pytz: 2023.3.post1
pywin32: 305
pyzmq: 25.1.2
qtconsole: 5.4.3
QtPy: 2.4.1
RandomWords: 0.4.0
requests: 2.31.0
scipy: 1.11.1
setuptools: 67.4.0
sfftk-rw: 0.7.3
six: 1.16.0
snowballstemmer: 2.2.0
sortedcontainers: 2.4.0
soupsieve: 2.5
sphinx: 6.1.3
sphinx-autodoc-typehints: 1.22
sphinxcontrib-applehelp: 1.0.8
sphinxcontrib-blockdiag: 3.0.0
sphinxcontrib-devhelp: 1.0.6
sphinxcontrib-htmlhelp: 2.0.5
sphinxcontrib-jsmath: 1.0.1
sphinxcontrib-qthelp: 1.0.7
sphinxcontrib-serializinghtml: 1.1.10
stack-data: 0.6.3
superqt: 0.5.0
tables: 3.8.0
tcia-utils: 1.5.1
tifffile: 2023.7.18
tinyarray: 1.2.4
tomli: 2.0.1
tornado: 6.4
traitlets: 5.9.0
typing-extensions: 4.9.0
tzdata: 2023.4
urllib3: 2.1.0
wcwidth: 0.2.13
webcolors: 1.12
wheel: 0.38.4
wheel-filename: 1.4.1
widgetsnbextension: 4.0.9
WMI: 1.5.1
File attachment: chimera problem.png
Attachments (2)
Change History (10)
by , 2 years ago
| Attachment: | chimera problem.png added |
|---|
comment:1 by , 2 years ago
| Cc: | added |
|---|---|
| Component: | Unassigned → UI |
| Owner: | set to |
| Platform: | → all |
| Project: | → ChimeraX |
| Status: | new → accepted |
| Summary: | ChimeraX bug report submission → Small checkboxes on second monitor |
comment:2 by , 2 years ago
| Status: | accepted → feedback |
|---|
Hi Jakob,
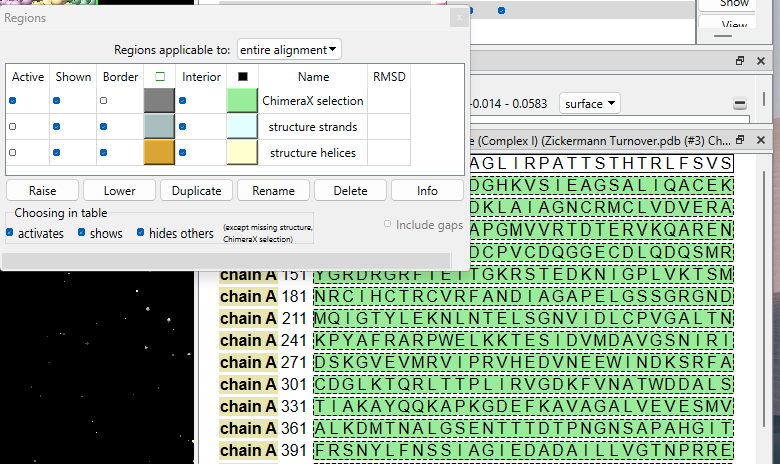
Thanks for reporting this problem. I'm hoping you can help me out with some more information. Try opening a structure (e.g. 7gt6) and show a chain sequence by clicking a link under the "Description" column in the chain table in the log. Bring up a context menu on the resulting sequence tool (usually by right-mouse click) and choose Annotations→Regions... in the context menu. In the Regions dialog that comes up, are the checkboxes in the table small? Are the checkboxes under the table (activates/shows/etc.) small?
--Eric
Eric Pettersen
UCSF Computer Graphics Lab
comment:3 by , 2 years ago
Hi Eric,
Thanks for your email. Looks like the boxes are small there too:
[cid:9e1fce87-7bb5-490f-a8c3-96e6b0d7059f]
Best
Jakob
________________________________
From: ChimeraX <ChimeraX-bugs-admin@cgl.ucsf.edu>
Sent: 08 February 2024 19:57:43
To: Jakob Andersson; pett@cgl.ucsf.edu
Cc: goddard@cgl.ucsf.edu
Subject: Re: [ChimeraX] #14554: Small checkboxes on second monitor
#14554: Small checkboxes on second monitor
----------------------------------------+----------------------
Reporter: jakob.andersson@… | Owner: pett
Type: defect | Status: feedback
Priority: normal | Milestone:
Component: UI | Version:
Resolution: | Keywords:
Blocked By: | Blocking:
Notify when closed: | Platform: all
Project: ChimeraX |
----------------------------------------+----------------------
Changes (by pett):
* status: accepted => feedback
Comment:
Hi Jakob,
Thanks for reporting this problem. I'm hoping you can help me out
with some more information. Try opening a structure (e.g. 7gt6) and show
a chain sequence by clicking a link under the "Description" column in the
chain table in the log. Bring up a context menu on the resulting sequence
tool (usually by right-mouse click) and choose Annotations→Regions... in
the context menu. In the Regions dialog that comes up, are the checkboxes
in the table small? Are the checkboxes under the table
(activates/shows/etc.) small?
--Eric
Eric Pettersen
UCSF Computer Graphics Lab
--
Ticket URL: <https://www.rbvi.ucsf.edu/trac/ChimeraX/ticket/14554#comment:2>
ChimeraX <https://www.rbvi.ucsf.edu/chimerax/>
ChimeraX Issue Tracker
comment:4 by , 2 years ago
| Status: | feedback → accepted |
|---|
comment:5 by , 2 years ago
| Cc: | added; removed |
|---|---|
| Owner: | changed from to |
| Status: | accepted → assigned |
Hi Jakob,
The fact that it is happening everywhere strongly implies it is a problem/bug in the Qt windowing toolkit that ChimeraX uses. Could you try the current ChimeraX daily build and see if it also has this problem? The daily build is using a significantly newer version of the Qt toolkit than the 1.7.1 release is, so there is some chance that the newer versions fixes this problem. Let us know what happens.
comment:6 by , 2 years ago
This is surely a bug in the Qt window toolkit.
The ChimeraX daily build is currently using the same Qt version as ChimeraX 1.7.1 and 1.7, namely Qt 6.3. ChimeraX 1.6 uses Qt 6.4.
We hope to soon update the ChimeraX daily build to Qt 6.6.2 but the release of that Qt has been delayed (it was supposed to be released 2 days ago). ChimeraX 1.7 uses the older Qt 6.3 because Qt 6.4 caused many crashes on Mac. Qt 6.6.1 caused different crashes on Mac that are supposed to be fixed in Qt 6.6.2.
The Qt window toolkit is riddled with bugs. You are fortunate it is only small checkbuttons.
The next step will be to try the ChimeraX daily build when we update it to Qt 6.6.2. That should happen in the next couple weeks after Qt 6.6.2 comes out. I will post a message to this ticket when ChimeraX is updated so you can test.
comment:7 by , 2 years ago
| Cc: | added; removed |
|---|---|
| Owner: | changed from to |
The ChimeraX daily build is now using Qt 6.6.2 while ChimeraX 1.7.1 uses the much older Qt 6.3. You could try the ChimeraX daily build and see if it still has the small checkbox problem.
If that does not fix it there is probably nothing we can do.
Reassigning to Eric in case the Models tool and Region pane do some quirky style sheet settings that may be causing the small checkbuttons.
comment:8 by , 2 years ago
| Resolution: | → limitation |
|---|---|
| Status: | assigned → closed |
There is no stylesheet tomfoolery going on.
Added by email2trac