Opened 8 years ago
Closed 8 years ago
#1126 closed defect (duplicate)
Functionality 'nucleotides' not working
| Reported by: | Owned by: | Greg Couch | |
|---|---|---|---|
| Priority: | normal | Milestone: | |
| Component: | Depiction | Version: | |
| Keywords: | Cc: | ||
| Blocked By: | Blocking: | ||
| Notify when closed: | Platform: | all | |
| Project: | ChimeraX |
Description
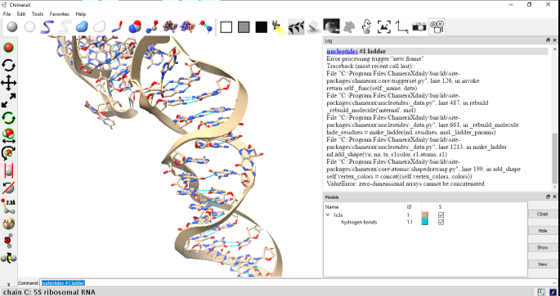
Hi everybody, I’m not able to represent the side chains of an RNA molecule as a ladder with the command ’nucleotides’. I’ve tried it in ChimeraX 0.6 and ChimeraX 0.7 daily and it is simply not represented. In the image below I’m testing the command on a 5S ribosomal RNA and I got the error of an empty Array. Sometimes I don’t get any error, the command seems to be recognized (in the log) but nothing changes on the molecule itself. I also tried to show all atoms, hide atoms, hide ribbon, delete hbonds, look for hbonds again, specify the model (#1) only, specificy the hydrogen bonds (#1.1) but nothing seems to work. Am I doing something wrong? Thank you for your consideration. André Rosa [cid:image002.png@01D3ED19.13D3A880]
Attachments (1)
Change History (3)
by , 8 years ago
| Attachment: | CB1900E4FF5B4986A3EBB73562E9E096.png added |
|---|
comment:1 by , 8 years ago
| Component: | Unassigned → Depiction |
|---|---|
| Owner: | set to |
| Platform: | → all |
| Project: | → ChimeraX |
| Status: | new → assigned |
comment:2 by , 8 years ago
| Resolution: | → duplicate |
|---|---|
| Status: | assigned → closed |
Apparent duplicate of #1125
Note:
See TracTickets
for help on using tickets.
Added by email2trac