Axes, planes, and centroids can be defined from sets of atoms using the
Axes/Planes/Centroids tool
or the command
define.
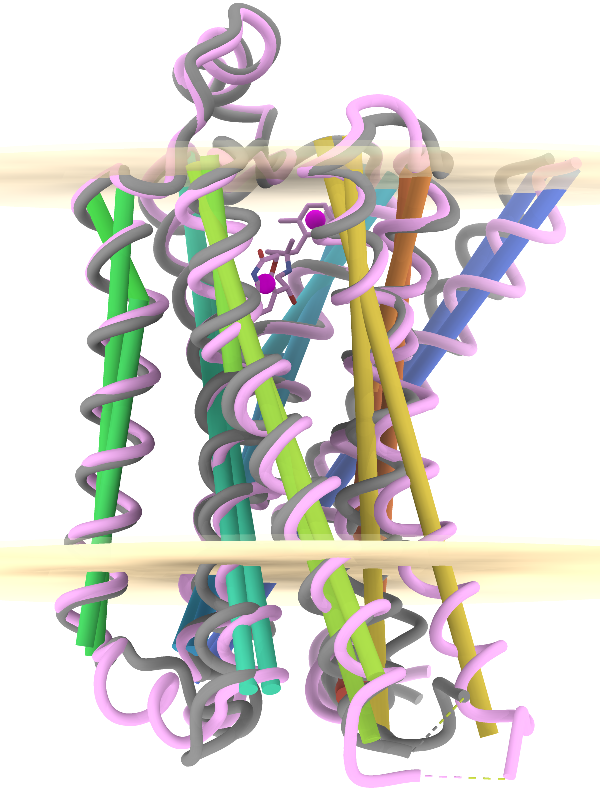
The image shows the β2-adrenergic receptor
in inactive (gray) and activated (pink) conformations,
PDB 2rh1
and 4lde
respectively. Hydrophobic membrane boundaries (light tan) are from the
Orientations of Proteins in Membranes (OPM) database.
Plane objects were calculated from the membrane-boundary pseudoatoms in
4lde-OPM.pdb.
Axes were calculated for the α-helices in both structures
and rainbow-color-coded from blue at the N-terminus to red at the
C-terminus. Centroids (magenta) were calculated from the two ring systems
of the agonist in the activated structure.
For image setup other than position, see the command file
axes-gpcrs.cxc.
Distances and angles between the defined objects can be measured using the
Axes/Planes/Centroids tool
or commands, for example, to reveal that the ring systems of the agonist
are about 9 Å apart and 3.3 and 8.7 Å from the outer membrane
boundary; that upon activation, TM6 (gold axes) tilts by about 15°
away from the helix bundle; and that TM4 (green axes, left side of image) makes
an angle of about 80-85° with the membrane (see
axes-measurements.cxc).
More features...