Preferences File
A Chimera preferences file contains settings from the
Preferences dialog and additional information
such as recently accessed files and directories.
A particular preferences file can be specified
with --preferences
at startup. Otherwise,
Chimera will look for a file named preferences
first in the current directory (the startup location, unless
cd has been used), if not found
then in a .chimera subdirectory of the current directory, and if
not found in either of those places, then:
- on Mac or Linux, in the .chimera subdirectory of the
user's home directory
- on Windows 10, in
C:\Users\username\AppData\Roaming\.chimera\
(possibly different for network accounts)
- on some older Windows versions, in
C:\Documents and Settings\username\Application Data\.chimera\
The file location is reported in the
Preferences preferences.
Preference changes and other information from the current session
will be saved to this file unless it has been designated
read-only.
This system allows a user to maintain a single set of preferences
for all Chimera sessions, or different preferences specific to data in
different directories, or a combination of these.
If no preferences file is found,
Chimera will look for a writable location for a new preferences file
in the following order: the user's home directory
(a subdirectory .chimera with a preferences file within it
will be created), a subdirectory .chimera of the current directory,
the current directory. If none of these directories are writable,
an error message will appear.
Preferences Dialog
Choosing Favorites... Preferences from the menu opens the
Preferences dialog.
Options within are grouped by Category:
For just the category being shown:
Reset replaces the current settings with the original defaults,
Restore replaces the current settings with
those previously saved in the
preferences file,
and Save saves the current settings to the
preferences file.
Equivalent operations can be performed for
all categories at once
in the Preferences section
of the Preferences dialog.
Help opens this manual page in a browser window, and
Close dismisses the Preferences dialog.
Default settings are indicated below in bold.
The Animation section of the
Preferences dialog is only available after a
scene
has been saved or the
Animation tool started.
See also: scene
- Scene name prefix (default nothing)
- text to prepend to the integer names of scenes saved with the
Animation dialog
- Scene thumbnail size (default 48 pixels)
- width and height of scene thumbnails in the
Animation
and Rapid Access interfaces
- Transition name prefix (default tr)
- Warn about model closure (true/false)
- whether to show a warning and allow cancellation when the user attempts
to close a model that is included in a saved scene
The Background section of the
Preferences dialog includes settings for the
graphics window background.
See also: background
- Background method (solid/gradient/image)
- whether the background should be a single solid color (default),
a gradient of multiple colors, or an image
- Background color (a
color well, default No Color,
equivalent to black)
- color to use when the
background method is solid
- Background gradient
(a palette well,
default HLS-interpolated white to blue)
- palette to use when the
background method is gradient;
left-to-right order in the palette well corresponds to bottom-to-top order
in the graphics window
- opacity (default 1.0)
- gradient opacity, ranging from 0.0 (completely transparent) to
1.0 (completely opaque); transparency reveals
the current solid background color.
If a transparent gradient is used together with a
transparent background, the background of
an image saved from Chimera as PNG or TIFF will not only show the blending
of the gradient over the solid color, but also have the same overall
opacity as was specified for the gradient.
- Background image (an image well, default No image)
- image to use when the
background method is image;
clicking the well brings up a file browser for locating and opening
an image file, where accepted formats include PNG, TIFF, and JPEG.
The image can be fit into the graphics window in any of the following ways:
- zoomed (default)
- automatically scaled to fill the window, top/bottom or sides
cropped as needed (no image distortion)
- stretched
- automatically scaled to fill the window, stretched or squashed as needed
- tiled - tiled to fill the window
- centered
- centered with any surrounding border shown in the current
solid background color
Additional image settings:
- scale (default 1.0) - image scale factor,
applies only when the image is tiled or centered
- opacity (default 1.0)
- image opacity, ranging from 0.0 (completely transparent) to
1.0 (completely opaque); transparency reveals
the current solid background color.
If a transparent background image is used together with a
transparent background, the background of
an image saved from Chimera as PNG or TIFF will not only show the blending
of the background image over the solid color, but also have the same overall
opacity as was specified for the background image.
The Command Line section of the
Preferences dialog is only available after the
Command Line has been shown.
The Fetch section of the
Preferences dialog controls handling of data
retrieved over the Web from various databases. See also:
open,
PDB preferences,
Fetch by ID
- Save fetched files (true/false)
- whether to save fetched files locally; even if this option is true,
no files will be saved unless a download directory has also been specified
- Use local files (true/false)
- whether to look in the download directory and use a previously fetched
copy (if any) when Fetch by ID or
the open command is used
to fetch a file from a database; when true, can be overruled by using the
Fetch by ID option to
Ignore any cached data
- Download directory (default ~/Downloads/Chimera)
- where to store fetched files locally; subdirectories for the different
databases will be created as needed. On Windows XP, the default is
~/My Documents/Downloads. If the specified location does not exist,
fetched files will not be cached.
The General section of the
Preferences dialog includes:
- Confirm quit (always/conditional/never)
- when to ask for confirmation before exiting Chimera;
if conditional, only when certain tools detect unsaved work.
If always or conditional, a conditional check will
also apply to closing a session.
Regardless of this setting, confirmation will not be requested when the
command stop is encountered within a
command file.
- File lists path style (file - leading path/full path) -
how to list files and their locations in certain dialogs (such as
Files to read at startup within the
Command Line preferences);
a change will not be evident until after Chimera is restarted
- Fullscreen graphics (true/false) - whether the graphics
window is in fullscreen mode
(available in Windows, Linux, and Mac-X11 versions of Chimera)
- Try to avoid placing dialogs over main window (true/false)
- whether to try to place additional Chimera dialogs alongside the
graphics window instead of on top of it
- File browser confirms overwrite (true/false)
- whether to ask for confirmation before overwriting a file with
a save dialog
- Use short open/save browser (true/false) - whether
open/save dialogs
should show only the two lowest directory levels
(more details)
- Open dialog starts in directory from last session
(true/false) - whether the file browser
most recent location
should be remembered between sessions
(default true on Windows and Mac, false on Linux).
- Debug OpenGL on startup (true/false) - whether to open
Debug Graphics
Driver at startup to selectively enable/disable OpenGL
features before starting Chimera.
This should be used only to find workarounds for
graphics driver bugs,
because in the absence of such problems, changing the settings from the
system defaults is expected to degrade Chimera performance and/or appearance.
Debug Graphics
Driver can also be called with the startup option
--debug-opengl.
- Update check interval
(never/daily/weekly/biweekly/monthly/quarterly)
- how often to check whether a newer version of Chimera
(snapshot or production release) is available
- Initial window size
(remember last/fixed)
- how to determine the size of the window at startup
- Fixed size - pixel width and height for fixed
initial window size
In the Mac version of Chimera only:
- Menus in windows on Mac (true/false)
- whether to show Chimera menus in-window as well as across the top
of the screen
The Image Credits section of the
Preferences dialog specifies information to be included
in saved image files (TIFF, PNG).
This information does not affect the image itself.
- Artist (user's login name by default) - optional, name of the
image creator
- Copyright (blank by default) - optional, a copyright statement
The Labels section of the Preferences
dialog contains settings for 3D labels (those which move along with
the structure) and pop-up atomspec balloons. The method of
residue label positioning
can be set in the
molecule model attributes panel
or with the command setattr.
See also:
2D Labels
- Label font (Sans Serif/Serif/Fixed)
(Normal/Bold/Italic/Bold Italic) (6/8/9/10/11/12/16/18/24/30/36)
- font typeface, style, and size to use for 3D labels
- Labels on top (true/false)
- whether to draw 3D labels in front regardless of their Z-offsets
- Show atomspec balloon
(true/false) -
when the cursor is paused over an object (atom, bond,
surface piece, etc.),
whether to show the corresponding label information in a balloon
in the graphics window, and PDB chain information, if available,
in the status line.
The number of decimal places shown for bond distances can be set with the
Distances tool.
- Atomspec display style
(simple/command-line specifier/serial number)
- how to list atoms in atomspec balloons,
Structure Measurements, and
Metal Geometry.
Atom naming styles:
- simple - residue name, residue specifier, and atom name
(for example, HIS 16.A ND1)
- command-line specifier - a
specification string that could be
used in the Command Line
(for example, :16.A@ND1)
- serial number - atom serial number (for example, 126)
Model number (for example, #0)
will also be included when multiple models are present.
The Messages section of the
Preferences dialog controls the disposition of various
messages and whether balloon help is shown.
- Show status line (true/false) - whether to show a
line for status messages
under the main Chimera window (otherwise, status messages are not visible);
can also be set from the
Accelerators dialog
- Clear status line after (5 seconds/10 seconds/20 seconds/30
seconds/1 minute/never) - how long a status message will be shown in
the status line (when not overwritten by subsequent status messages);
applies to status messages appearing in certain dialogs as well as
the main Chimera status line
- Show balloon help (true/false) - whether to show
balloon help,
explanatory text that pops up when the cursor is left in certain positions
- Command (reply log only/dialog) - whether command messages
should appear in the Reply Log
only, or also generate a dialog
- Warning (reply log only/dialog) - whether warnings
should appear in the Reply Log
only, or also generate a dialog
- Error (reply log only/dialog) - whether error messages
should appear in the Reply Log
only, or also generate a dialog
Mouse button functions
including the manipulation of models
can be reassigned using the Mouse section
of the Preferences dialog.
Functions may be assigned to the left, middle, and right mouse
buttons alone and in combination with the Ctrl key.
(Modifier keys allow accessing these functions with a
touchpad or simpler mouse.)
There can be no more than one function checked per row
(per button or Ctrl-button).
| icon | meaning |
default assignment |
 |
rotation |
1
(left button) |
 |
XY-translation |
2
(middle button) |
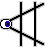 |
scaling |
3
(right button) |
 |
picking
(selection from screen) |
Ctrl-1 |
 |
Z-translation |
Ctrl-2 |
 |
click item
(atom, bond, surface vertex...)
to center, set as fixed
center of rotation;
click empty space to restore
front center mode
| Ctrl-3 |
 |
pop-up Chimera menu |
(none)
see alternatives |
 |
label-dragging
(XY, +Shift for Z) |
(none)
|
- Continue rotation after mousing (true/false) - whether
rotation should continue when the corresponding mouse button is released
while the cursor is in motion
- Use scrolling (true/false) - whether
mouse or touchpad scrolling should also scale the view.
On a Mac, it may be necessary to set the
multitouch preference to false
to make touchpad scrolling available.
In the Mac version of Chimera only:
Some tools create additional mouse modes, for example:
The New Molecules section of the
Preferences dialog controls how subsequently opened molecule models
will initially appear.
New Molecules settings can be circumvented by opening structures
with the command open noprefs;
this prevents inconsistent behavior of
command files and
demos
potentially caused by different preference settings of different users.
By default, these preferences are not applied to structures opened via
Chimera web data
(chimerax) files, but noprefs="false" can be set in the
chimerax file to indicate that the preferences should be applied.
Regardless of noprefs options, however, the preferences for
metal complex depiction and mol2 naming are always applied.
See also:
open,
molecule model attributes,
presets
- smart initial display (true/false) -
show subsequently opened molecule models with settings similar to the
ribbons preset,
but without rainbow-coloring chains or altering global parameters
such as background color.
Several other New Molecules preferences are overridden and grayed out
when smart initial display is set to true:
- ribbon display (off/on)
- whether to show ribbon; ribbons are only drawn for proteins and nucleic acids.
Protein helix and strand assignments are taken from the input structure
file or generated with ksdssp.
- ribbon cross section (edged/flat/rounded/any additional
styles
made with the
Ribbon
Style Editor)
(see also ribrepr)
- ribbon scaling (Chimera default/licorice/any additional
scalings
made with the
Ribbon
Style Editor)
(see also ribscale)
- ribbon hides backbone atoms (true/false) -
whether ribbon display hides backbone atoms for the corresponding residues
(see the command
ribbackbone for details)
- show atoms (true/false)
- whether to display all atoms/bonds (except backbone when hidden by ribbon)
- per-atom coloring (none/by element/by heteroatom)
- whether to leave atom colors unassigned (none;
atoms will inherit model colors)
or to assign element colors
to all atoms (by element) or only the non-carbon atoms
(by heteroatom)
- atom style (dot/ball/endcap/sphere) - atom
draw mode
(excluding monatomic ions)
- bond style (wire/stick) - bond
draw mode
- auto-chaining (off/on)
- whether to connect atoms that precede and follow undisplayed segments
(whether to draw pseudobonds between them)
-
use new color for each model (true/false) -
set the model-level color
(see coloring hierarchy)
differently for each new molecule model;
submodels with
the same main model number will be assigned the same model-level color.
This setting can also be controlled with
set/~set autoColor.
If the solid background color is
black or white, automatic model colors are
0:tan, 1:sky blue, 2:plum, 3:light green,
4:salmon, 5:light gray, 6:deep pink, 7:gold,
8:dodger blue and 9:purple
(see named colors):
| #0 |
#1 |
#2 |
#3 |
#4 |
#5 |
#6 |
#7 |
#8 | #9 |
Thereafter, or for all models if the solid background color
is not black or white, an algorithm is used to produce colors
distinguishable from the background and from other models.
- otherwise, use color (a
color well, by default
a light gray) - model-level color if not using a new color for each model
- stick scale (default 1.0)
- scale factor for bonds in the stick
draw mode.
The stick scale is multiplied by individual bond radii (default
0.2 angstroms) to generate stick radii in angstroms.
Changing stick scale is preferable to changing all of the bond radii
in a model, because the former will also scale singleton atoms in the
endcap draw mode appropriately. Either way,
the other endcap atoms (those participating in bonds) will be scaled
to match the thickest of the attached bonds.
- ball scale (default 0.25)
- scale factor for atoms in the ball
draw mode.
The ball scale is multiplied by individual atom
VDW radii to generate ball radii in angstroms.
- line width (default 1.0)
- pixel width of lines depicting bonds (when in the wire draw mode)
- monatomic ion style
(dot/ball/endcap/sphere) - draw mode
to use for monatomic ions
- metal complex representation
(dashed lines/solid lines/springs)
- how to show metal coordination pseudobonds
- metal complex color (a
color well, medium purple
by default) - color to use for
metal coordination pseudobonds
- metal complex line width (default 2.0)
- pixel width of lines representing metal coordination
pseudobonds
- Mol2 model naming
(file name/Mol2 molecule name/Mol2 molecule comment) -
how to name models read from Mol2 format: by file name, by molecule name
given within the file (on the line below @<TRIPOS>MOLECULE), or
by comment text (the sixth line below @<TRIPOS>MOLECULE)
The New Surfaces section of the
Preferences dialog sets parameters for subsequently created
molecular surfaces.
See also:
surface,
molecular surface attributes,
presets
- representation
(solid/mesh/dots) - display type
- probe radius (default 1.4) - radius
in Å of the probe sphere used to compute the surface. A larger probe
decreases surface bumpiness because it fits into fewer crevices.
A radius of 1.4 Å is commonly used to approximate a water molecule.
- vertex density (default 2.0)
- vertices per Å2.
Greater density results in a smoother surface but
increases computational demands for calculating and moving the surface.
- line width (default 1.0) - pixel width of lines
used in the mesh surface representation
- dot size (default 1.0) - pixel size of dots
used in the dot surface representation
- show disjoint surfaces (true/false)
- whether to check for multiple disconnected parts of a surface
(components)
rather than assuming there is only one; increases calculation time
The PDB section of the
Preferences dialog includes:
- Fetch from web as necessary (true/false) -
whether to retrieve a file specified by PDB ID from the
Protein Data Bank website
when it is not found locally (see the Web Access
preferences for proxy settings, if needed).
Chimera first looks for a local installation of the Protein Data Bank,
then in any personal PDB directories,
and then in the fetch download directory (unless
disallowed in the Fetch preferences) before
resorting to a web fetch. The default places to look for a local installation
are /usr/mol/pdb/ and then /mol/pdb/
— these can be changed by editing the Python file
share/chimera/pdbDir within a Chimera installation.
- Personal PDB directories
- additional directories in which to look for PDB files.
Directories can be added to or deleted from the list.
The directory structure can be flat, where
a single directory contains files named xxxx.pdb (and xxxx is the
PDB ID), or hierarchical,
with pathnames of the form xy/pdb1xyz.ent, where 1xyz is the
PDB ID and subdirectory names are the
two middle characters of the IDs. PDB filenames of the form
pdb1xyz.ent can also be used in the flat directory structure.
The POV-Ray Options section of the
Preferences dialog sets specifications for
raytracing with POV-Ray.
- POV-Ray executable [ location ]
- the location of the POV-Ray executable. This is filled in automatically
for the executable included with Chimera, but another location
could be specified.
- Show preview (true/false) - whether to show a preview;
regardless of this setting, however, a preview will not be shown
during movie recording or
when Chimera is in nogui mode.
- Quality (default 9, range 0-11)
- output image quality, adjusted by omitting calculations that are normally
performed. For example, values < 4 yield images without shadows
[quality details at the POV-Ray site].
- Antialias (true/false)
- whether multiple samples should be averaged to produce a single output pixel;
otherwise, POV-Ray will simply trace one ray per pixel.
Analogous to (but independent of)
Chimera image supersampling,
this supersampling in POV-Ray smooths edges in output images
[antialias details at the POV-Ray site].
- Antialias method
(fixed/recursive) - how pixels should be supersampled when
antialiasing is used.
In the fixed method, POV-Ray initially traces one ray per pixel.
Each pixel is compared to its neighbors to the left and above.
If the colors of two pixels differ by more than the
antialias threshold, each of the pixels
will be subsampled according to the antialias depth
(if the depth is 3, a grid of 3*3=9 samples will be taken per output pixel).
In the recursive method, POV-Ray initially traces 4 rays at the corners
of each pixel. Where the colors of adjacent samples differ by more than the
threshold, subsamples will be taken and compared.
This process is repeated until adjacent colors agree within the threshold,
or the maximum number of subdivision cycles
(the specified antialias depth) has been performed.
- Antialias depth (default 3, range 1-9)
- meaning depends on which antialias method is used
- Antialias threshold
(default 1.0, range 0.0-3.0)
- two colors are compared by summing the
absolute differences in their red, green, and blue components;
differences greater than the threshold trigger further sampling
when antialiasing is used.
Lower threshold values increase the number of samples and the calculation time,
whereas a threshold of 3.0 will suppress further sampling.
- Jitter (true/false)
- whether to introduce noise by randomly wiggling the supersample locations
when antialiasing is used; not recommended for
movie recording, as the random noise
will make pixels vary and flicker from frame to frame
[jitter details at the POV-Ray site]
- Jitter amount (default 0.5)
- values are specified relative to 1.0, which is
the most jitter that can be performed
while retaining supersamples within the original pixel
- Transparent background
(true/false/same as chimera)
- whether to make the background transparent by including transparency
(alpha) values for output pixels
[output alpha details at the POV-Ray site].
The same as chimera option means to follow the current
transparent background
setting in the Save Image dialog.
It is also necessary to turn off
depth-cueing
to obtain background transparency in raytraced images.
- Wait for POV-Ray to finish (true/false)
- whether to prevent further commands from executing until
POV-Ray has finished generating the image.
During movie recording,
the behavior will always be as if this were set to true.
- Keep POV-Ray input files (true/false)
- whether the POV-Ray input files containing the scene (*.pov) and raytracing
options (*.ini) should be kept or deleted after the image has been rendered
The Preferences section of the
Preferences dialog includes:
- Preferences file - the location of the
preferences file for the current session
- Read-only - whether the
preferences file for the current session
should be treated as read-only.
The initial setting reflects the file permissions at Chimera startup.
When Read-only is false, Chimera may update the file
with changes in preference settings and other information;
switching to true prevents subsequent changes in the file.
When Read-only is true, Chimera will not update the file;
switching to false (if the file is already writable, or is owned
by the user) will enable its modification by Chimera.
- All categories:
The Presets section of the Preferences
dialog allows specifying Personal preset directories
in which Chimera should look for custom preset scripts.
Directories can be added to or deleted from the list.
Preset script file types:
Custom presets will appear in the Presets
menu in alphabetical order by name, which is determined from
script filename: name.preset.suffix
where suffix indicates the file type, as listed above.
In other words, the filename must end in “.preset.com,”
“.preset.cmd,” or “.preset.py.”
Custom presets can also be accessed with the
preset command.
The Selection section of the Preferences
dialog controls how selections are highlighted.
- Selection highlight method (outline/fill) - how
a selection is highlighted in the graphics window
- Selection highlight color (a
color well, green by default) -
what color to use for highlighting a selection
in the graphics window
The Tools section of the
Preferences dialog allows control over the following
Settings for each entry in the
Tools menu:
- Auto Start - whether to launch the tool automatically
upon Chimera startup
- On Toolbar - whether to include the tool icon in the
toolbar
- In Favorites - whether to include the tool in the
Favorites menu
- Confirm on Start - whether to ask
for confirmation before starting the tool
The toolbar is shown when
it includes at least one icon.
- Toolbar placement (left/right/top/bottom/Rapid Access)
- whether to put the toolbar along one side of the Chimera graphics window,
or only in the Rapid Access interface
The Third-party plugin locations section allows users to
specify additional directories in which Chimera should look
for tools. Directories can be added to or deleted from the list.
The Web Access section of the
Preferences dialog includes:
- Confirm open of commands or code (once per session/each
time/never) - whether/how often to
ask for
confirmation before opening certain types of
data
linked to web pages.
- Accept web data (true/false) - whether the running
instance of Chimera can be used as a helper application for
data
linked to web pages (and can receive files specified with
chimera --send).
On Mac, this preference is not shown, but is effectively
true unless Chimera was started from the terminal command line.
A browser must be
configured
to send the files to Chimera.
If there is no running instance of Chimera enabled to accept web data,
another instance of Chimera will be started and used to open the data.
If there are multiple running instances of Chimera set to
accept web data, the file will be sent to the instance that
most recently had focus (was most recently clicked into).
- Use HTTP proxy (true/false) - whether to use
a proxy to connect to the internet through a firewall
(for example, to fetch a PDB file from the
Protein Data Bank website);
the server and port number settings (below) should generally be the same as
those used for web browsers
- Proxy server - the hostname or IP address of the proxy server
- Proxy port - proxy port number to use
UCSF Computer Graphics Laboratory / May 2021