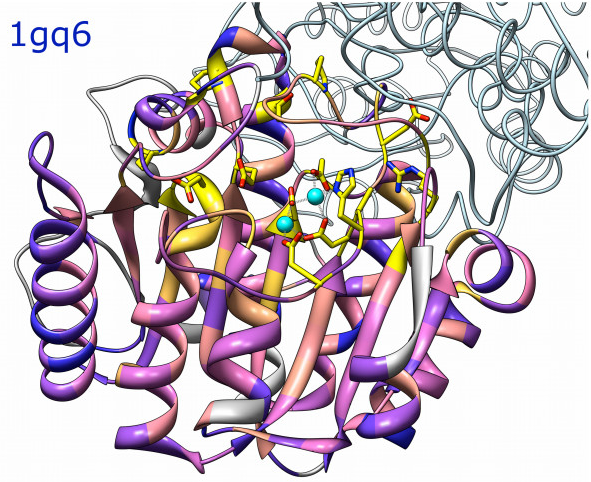

Atomic models and sequences
Morphing between conformations.
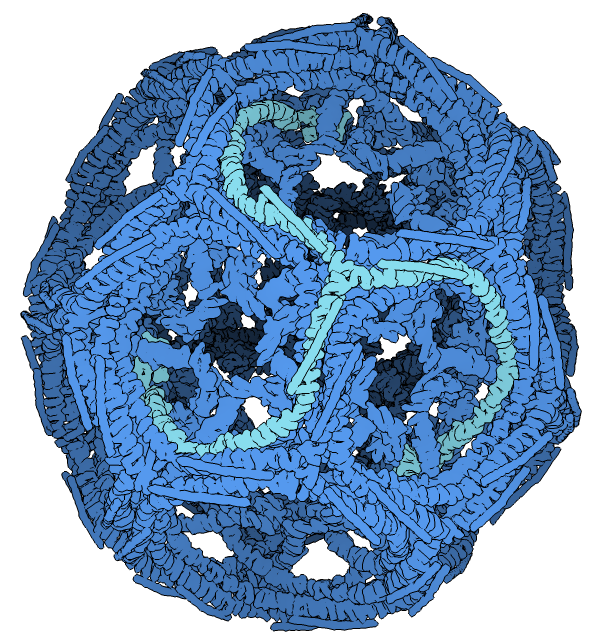
Molecular assemblies
Tom Goddard
January 23, 2009
  Atomic models and sequences |
Morphing between conformations. |

Molecular assemblies |
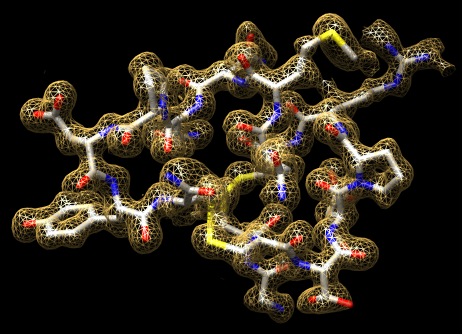
Crystallography maps |
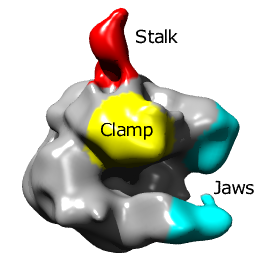
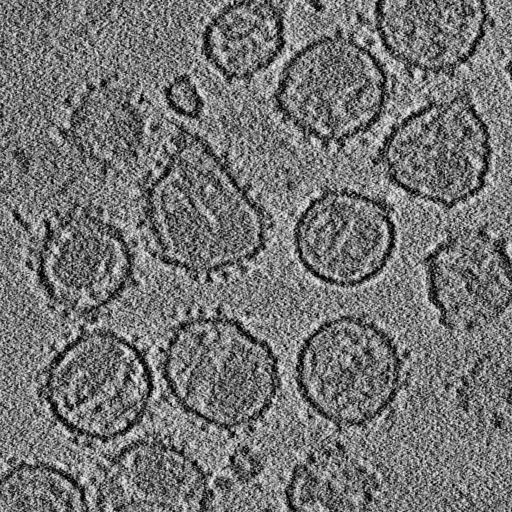
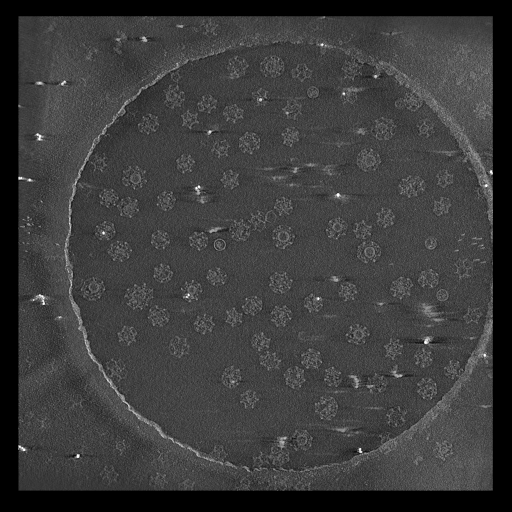
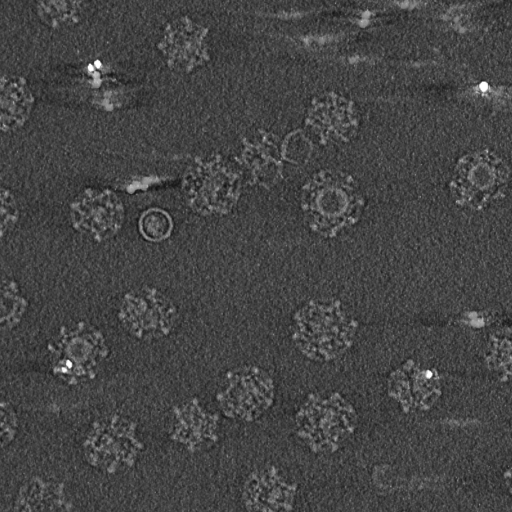
Single particle reconstructions |
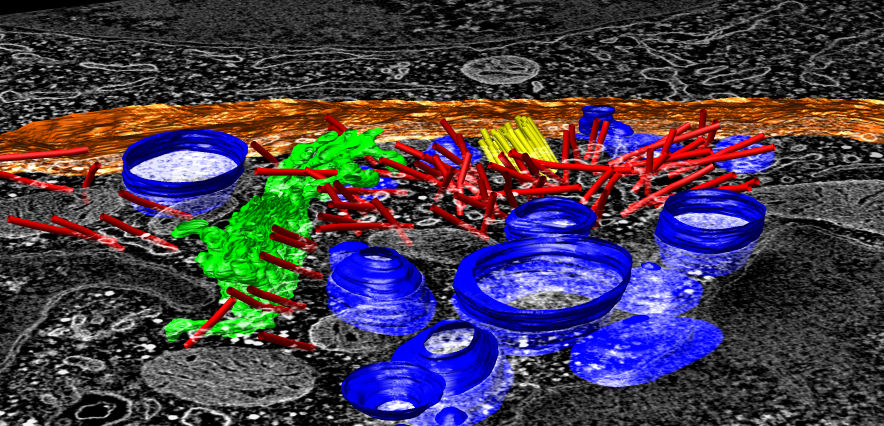
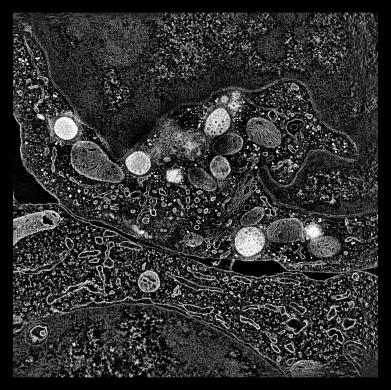
EM tomography |
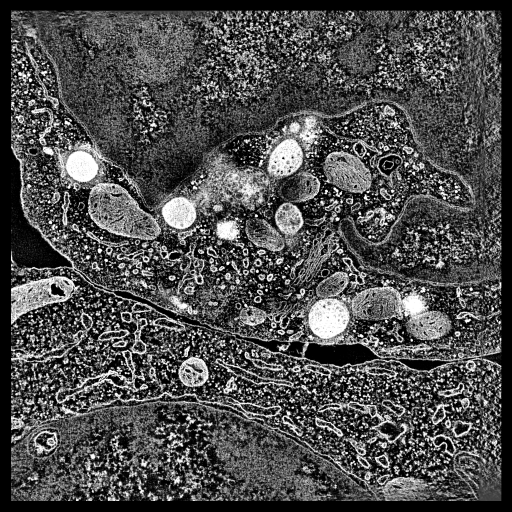
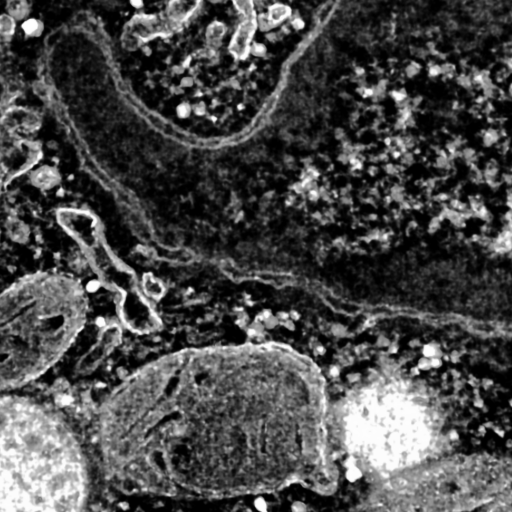
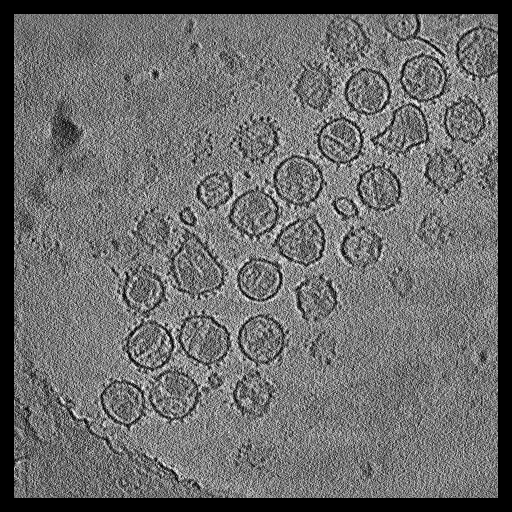
|
|

|

|

|
|
|
|

|

|
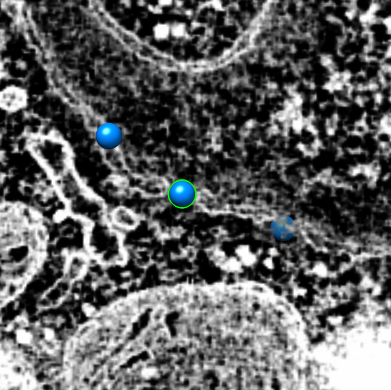
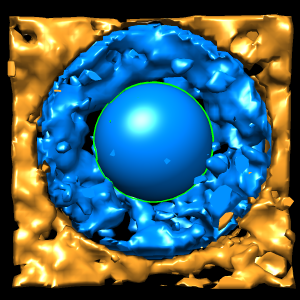
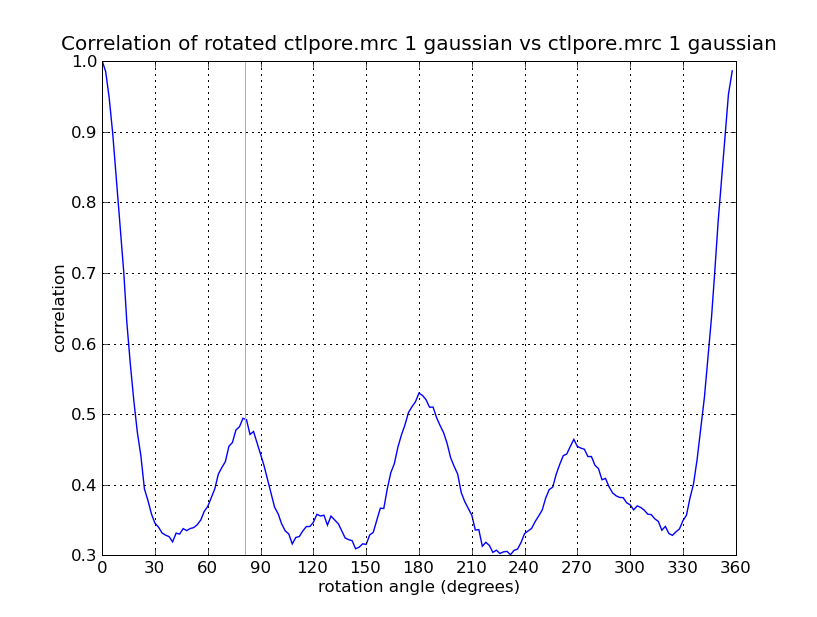
To extract density new many membrane embedded objects like 100 virus spikes rotating a box around each is time consuming. Better to extract density near membrane surface for the entire membrane.
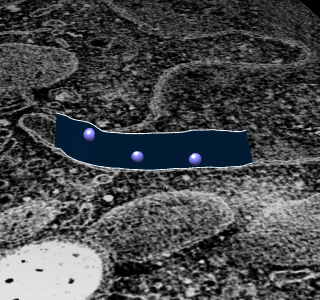
|
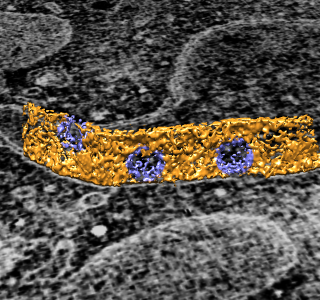
|
To show pores in their environment make a fly-through animation.
