Tool: Foldseek (Similar Structures)
The Foldseek tool (also called Similar Structures)
searches the PDB or
AlphaFold Database
for structures similar to a protein chain already open in ChimeraX.
The tool facilitates exploring up to hundreds of protein single-chain
structures by efficiently showing them in 3D as backbone traces,
potentially with ligands, and in 2D as sequence alignments or
reduced-dimensionality scatter plots based on backbone conformation.
-
The Foldseek search method
finds similar 3D structures (regardless of sequence similarity)
using extremely fast approaches that were developed for sequence searches.
It does so by describing the 3D interactions along a chain of amino acids
as a linear sequence of characters.
-
Alternatively, MMseqs2 (very fast) or
BLAST can be used to
search for protein structures by sequence similarity.
The tool can be started from the
Structure Analysis section of the Tools menu
and manipulated like other panels
(more...).
It is also implemented as the commands
foldseek,
similarstructures, and
sequence search.
See also:
AlphaFold,
ESMFold,
Blast Protein
Search Setup
Similar Structures List
Options
Sequence Plot and Residue Attributes
Traces
Cluster Plot
Ligands
References
[back to top: Foldseek]
Search Setup
The query should be chosen from the pulldown menu
of protein chains in structures already open in ChimeraX.
Choices of database to search:
Choices of search method:
- Foldseek – fast 3D structure search
using the Foldseek Search Service provided by the
Söding
and Steinegger
groups, with maximum number of hits 1000.
If this method is used, PDB refers to the
“pdb100” redundancy-filtered version of the PDB
created with Foldseek (one chain per cluster with 100% sequence identity
and ≥95% sequence overlap, reducing ~1 million chains to 340,000)
and Alphafold DB to the “afdb50” UniProt50 subset of the
database version 4.
- MMseqs2 – very fast sequence search
using the RCSB web service
- BLAST
– sequence search using the BLAST web service hosted by the
UCSF RBVI
Clicking Search sends the input parameters and structure to the
web service. When results are returned, a table
of similar structures is shown in the tool window.
[back to top: Foldseek]
Similar Structures List
Searching with the Foldseek (Similar Structures) tool
or the commands
foldseek,
sequence search, and
similarstructures
blast shows a table or list of hits in the tool window.
Because this list is relatively large, the ChimeraX graphics and/or
overall window may be resized; to avoid this, the tool can be
undocked from the main window
beforehand. See also the Tool windows start undocked setting in the
Window preferences.
Columns in the Similar Structures table:
- PDB – PDB-identifier_chain-identifier
– OR – (depending on which database was searched)
- AFDB
– UniProt accession number
- Identity – % sequence identity compared to the query
- E-value – significance value according to the search method
- % Close
– percentage of residues in the trimmed hit
that are close to the paired residue of the query (within the
Alignment pruning C-alpha atom distance,
default 2.0 Å)
- % Cover
– percentage of query residues paired with hit residues
- Species – source organism
- Description
– description of the protein or (if PDB)
the overall structure containing the protein
The % Close and % Cover values are only filled in automatically
by the Foldseek search method,
which uses and returns 3D coordinates.
For the other search methods (which are based on sequence only),
these columns can be filled in by using
similarstructures
fetchcoords to get α-carbon coordinates for the
corresponding structures.
One or more hits can be chosen (highlighted) in the table
by clicking and dragging with the left mouse button;
Ctrl-click (or command-click if using a Mac)
toggles whether a row is chosen.
Buttons across the bottom of the dialog:
- Open
– fetch the chosen
structures from the respective database
(Protein Data Bank
or AlphaFold Database) and process them
according to the options. For each structure,
the hit chain is superimposed onto the query chain by least-squares fitting
the α-carbons of the paired residues and iteratively pruning
far-apart pairs as described for the
align command.
- Sequences
– show an interactive heatmap of the sequence alignment of all hits
(details...)
- Traces
– show hit structures as thin tube backbones
(details...)
- Clusters
– show an interactive 2D scatter plot of the hits based on
their backbone conformations
(details...)
- Ligands
– show any solvent, ligands, and ions from the hit structures
(ligands...)
- Options
– show options for handling structures and
for setting whether buttons should apply to all hits or only the
chosen hits
- Help
– show this page in the Help Viewer
Choosing Save CSV or TSV File... from the tool's
context menu opens
a separate dialog for exporting the data (all rows and columns) to a
comma-separated or tab-separated values file, retaining the current sort order.
The current contents of the tool are also saved in
ChimeraX sessions.
In addition, search results are saved in ~/Downloads/ChimeraX
under subdirectories Foldseek, MMseqs2, or BLAST, with filenames
based on the query name, the database searched, and the search method,
ending with the suffix .sms.
These similar structures files
use a JSON file format specific to ChimeraX and are listed in the
File History for easy access.
Simply opening an .sms file loads the set of results into the
Similar Structures interface.
Doing another search or opening a file of previously saved results replaces the
contents of the Similar Structures table, since (currently) the tool
only allows showing one set of results at a time.
Sets of results are assigned names such as fs1, fs2, mm1, mm2, bl1, and bl2
that can be used in analysis commands even if the corresponding results
are not shown. However, the only way to get a set of results that is open
but not shown in the table is to use the showTable false option of
the search command.
The names of currently open sets can be listed with the command
similarstructures
list.
[back to top: Foldseek]
Options
Clicking Options shows/hides the following settings:
- Trim – delete any/all (default) of the following from
the retrieved structure:
- extra chains – for PDB entries, chains other than
the hit chain
- sequence ends – N- and C-terminal segments of the hit chain
that were not included in the sequence alignment returned by the search method
- far ligands – ligands, solvent, and ions > 3 Å
from the hit chain
- Alignment pruning C-alpha atom distance (default 2.0 Å)
– iterate the fit over the sequence-aligned pairs of CA atoms
so that only pairs within the specified distance are used in the final fit,
as described for the align command
- Traces, clusters and ligands for selected rows only
– whether the Traces, Clusters, and Ligands
buttons should show data for the
chosen hits only; otherwise,
show data for all hits regardless of which are chosen
[back to top: Foldseek]
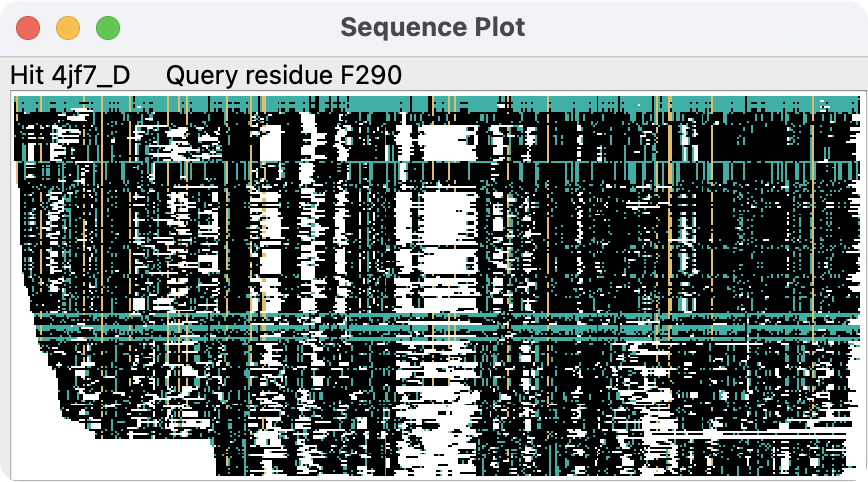
Sequence Plot and Residue Attributes
Clicking the Sequences button displays a high-level (without
amino acid codes) plot of the sequence alignment of all of the hits to
the query. The plot gives an overview of which parts of the query sequence
are matched by the hits, and the depth of coverage.
Each row of the image is one sequence, so 200 hits would
produce an image 200 pixels tall. The columns of the image correspond
to the residues of the query structure. Initially, pixels in the plot are
colored as follows:
-
– no aligned residue
-
– residue of the same amino acid type as the query
in a column where ≥0.5 of the residues have that same type
(and the column contains at least 10 residues)
-
– residue of the same amino acid type as the query
but not meeting the column criteria above
-
– residue of a different amino acid type than the query
Different coloring can be applied with the
similarstructures
sequences command.
Hovering the mouse over the sequence plot shows pop-up labels to indicate
the underlying row (hit structure) and column (query residue number).
Left- or right-clicking the plot raises a context menu,
in which some entries reflect the row or column position of the click:
- Open structure [hit]
– fetch the structure and superimpose it on the query
as described for the Open button
- Show [hit] in table
– scroll the table of results to the corresponding row
- Select query residue [query residue]
– select the corresponding residue in the query
- Order sequences by:
- e-value – lowest to highest E-value (default,
if clustering by coverage only gives one cluster, see below)
- cluster – grouping the sequences by which part of the
query they cover (default, if clustering by coverage gives >1 cluster)
- identity
– percent sequence identity compared to the query
- mean LDDT
– average local distance difference test (LDDT)
over all residues in a hit structure.
The LDDT indicates the similarity of a hit residue to the aligned
query residue in a neighborhood of 15 Å from the query residue
α-carbon
(see Mariani et al.,
Bioinformatics 29:2722 (2013)).
- Color conserved (default)
– by conservation of the query residue type
as described above
- Color by LDDT
– by LDDT of each aligned residue in each structure:
(if both of the above Color... options are turned on, only the
positions that would otherwise be shown in black are colored by
LDDT instead)
- Color query structure by:
- coverage
– number of residues (non-gap characters) in the aligned column
(query residue attribute coverage):
where N is the number in the most highly populated column of the alignment
- conservation
– fraction of hits in the aligned column with the most prevalent
residue type in that column, not necessarily the same residue type
as in the query
(query residue attribute conservation):
- highly conserved
– red where
conservation ≥ 0.5, otherwise gray
- local alignment
– average LDDT at that position across all
aligned structures
(query residue attribute lddt):
- Save image – save the sequence plot as a PNG image file
The query residue attributes
coverage, conservation, and lddt
are assigned when the sequence plot is shown.
Different coloring schemes to show these
attributes
can be applied to the query structure with
Render by Attribute or the command
color byattribute.
[back to top: Foldseek]
Traces
Clicking the Traces button displays hit structures as
“licorice” (spaghetti-like) ribbons superimposed on the query,
for either all hits or just the chosen ones,
as per the options.
These traces are meant to give an overview of the variability
of a large number of stuctures and their coverage of the query,
and soft lighting
is recommended to better reveal their shapes.
Only backbone α-carbons are included in the condensed structural
information returned by a Foldseek search, not secondary structure
information, so the ribbons do not vary in style to show helix and strand.
MMseqs2 and Blast search results do not automatically
include α-carbon coordinates, but clicking the
Traces button will raise a dialog asking the user whether to
fetch them, since it may take several minutes to do so.
All of the hit structure α-carbons are loaded as a single atomic model,
one chain per structure, with chain ID set to the database ID of the structure.
The residue types of the hit are retained, but the residues are renumbered
according to the paired residues of the query structure.
The traces are initially displayed as follows:
- the ribbon is broken into segments where
two consecutive aligned α-carbons are >5 Å apart
- ribbons are shown for ≥5 contiguous α-carbons within a segment
and within 4 Å of the corresponding query α-carbons
- ribbons are shown for entire segments in which every α-carbon
is within 10 Å of its counterpart
Different parameters can be applied with the
similarstructures
traces command.
Ctrl-double-clicking a trace shows a
selection context menu
for the corresponding hit, with entries including:
- Open similar structure [hit]
– fetch the structure and superimpose it on the query
as described for the Open button
- Show [hit] in similar structures table
– scroll the table of results to the corresponding row
- Show all traces – show trace ribbons for all hits
- Show full traces – show entire traces
regardless of the distance and length criteria for initial display
- Show only close traces – go back to showing
only the parts that meet the distance and length criteria for initial display
- Show only trace [hit]
The trace ribbons can be shown/hidden or colored selectively with the
menu above and the cluster plot context menu.
[back to top: Foldseek]
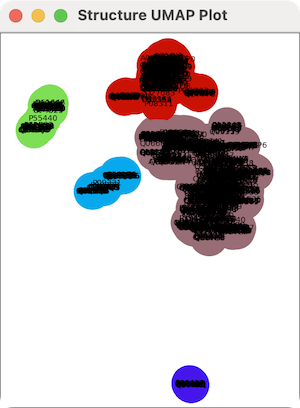
Cluster Plot
Clicking the Clusters button displays a scatter plot of the hits
clustered by backbone conformation, for either all hits or just the
chosen ones, as per the
options. Each structure is represented by a circle
labeled with its name. Clicking the button generates the plot as follows:
- the five residues in the query most conserved in the sequence
alignment of hits are identified and their α-carbons used as the
reference atoms
- for each hit,
the α-carbon (x,y,z) coordinates of the corresponding five residues
are concatenated to give a vector of length 15; hit structures
without a residue in any of the five alignment columns are omitted
- the vector is projected to a point in two dimensions with UMAP
(Uniform Manifold Approximation and Projection)
- the points in 2D are clustered by distance,
and the clusters are assigned random colors
Different parameters such as a different number of reference residues
can be specified with the
similarstructures
cluster command.
The plot can be zoomed by scrolling and translated with the middle mouse
button or trackpad equivalent. Clicking the plot raises a context menu.
Menu items acting on traces will generate them as needed (if not already
present) as described above, and those referring to
a specific hit only appear when the click is on a circle:
- Show traces for cluster [hit]
– show traces for all hits in the same cluster as the clicked one
- Show only traces for cluster [hit]
– show traces for all hits in the same cluster as the clicked one,
hide all other traces
- Hide traces for cluster [hit]
– hide traces for all hits in the same cluster as the clicked one
- Show all traces
- Show one trace per cluster
– per cluster, show only the trace of structure closest to the
average for that structure (minimum RMSD to average α-carbon positions)
- Show traces not on plot
– show traces for the hits that were omitted from the plot
due to not having a residue at one or more reference positions
- Hide traces not on plot
– hide traces for the hits that were omitted from the plot
due to not having a residue at one or more reference positions
- Color traces to match plot
- Change cluster [hit] color
– use system color editor to interactively change the color of all
circles for hits in the same cluster (or if coloring by species, from
the same species) as the clicked one
- Color by cluster (default)
- Color by species
– different random colors for different source species
- Show table row for [hit]
– scroll the table of results to the corresponding row
- Select rows for cluster [hit]
– choose table rows for all hits in the same
cluster (or if coloring by species, from the same species) as the clicked one,
and report their structure IDs in the Log
- Log structure names for cluster [hit]
– report structure IDs in the Log
for all hits in the same cluster as the clicked one
- Show reference atoms
– display the reference α-carbons as spheres
and select them
- Select reference atoms
– select the reference α-carbons
[back to top: Foldseek]
Ligands
Clicking the Ligands button copies the
ligands, ions, and solvent molecules (nonpolymer residues) from the hits
onto corresponding locations on the query structure,
for either all hits or just the chosen ones,
as per the options. A dialog will appear
to ask the user whether the structures should be fetched,
since it may take several minutes to do so.
Each ligand (ion, solvent) residue is evaluated for mapping onto the query
structure, as follows:
- protein residues within 5 Å of the ligand are identified
- if at least half of those nearby protein residues are paired with query
residues, the α-carbons of those pairs are fitted
- if the resulting RMSD is ≤3 Å, the ligand is copied to
corresponding position relative to the query structure
Different parameters can be applied with the
similarstructures
ligands command.
How many residues were copied and their residue types
are reported in the Log.
Often thousands of water molecules, and ions, and crystallization adjuvants
are found, and they can be hidden to get a better view of more interesting
ligands. For example, commands:
hide solvent
hide ions
hide :SO4
By default, the copied ligand, ion, and solvent residues are loaded as a
single atomic model, in which the chain ID of a residue is generated from the
PDB ID and chain ID of its source structure (e.g., 2cml_B).
Pausing the cursor over a residue in the graphics window shows
its name and chain ID in a pop-up balloon.
See also: AlphaFill
[back to top: Foldseek]
References
Foldseek.
The Foldseek method is described in:
Fast and accurate protein structure search with Foldseek.
van Kempen M, Kim SS, Tumescheit C, Mirdita M, Lee J, Gilchrist CLM, Söding J, Steinegger M.
Nat Biotechnol. 2024 Feb;42(2):243-246.
Many-against-Many sequence searching (MMseqs2).
The MMseqs2 method is described in:
MMseqs2 enables sensitive protein sequence searching for the analysis
of massive data sets.
Steinegger M, Söding J.
Nat Biotechnol. 2017 Nov;35(11):1026-1028.
MMseqs2 desktop and local web server app for fast, interactive sequence
searches.
Mirdita M, Steinegger M, Söding J.
Bioinformatics. 2019 Aug 15;35(16):2856-2858.
Local distance difference test (LDDT):
lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests.
Mariani V, Biasini M, Barbato A, Schwede T.
Bioinformatics. 2013 Nov 1;29(21):2722-8.
UCSF Resource for Biocomputing, Visualization, and Informatics /
November 2025