
February 27, 2007
This script generates images for a set of density maps. It was written to create virus images from electron microscopy maps for the Virus Particle Explorer web site (VIPERdb, viperdb.scripps.edu).
Before running the script viper_em_images.py you need to edit it to specify the directory containing maps, the directory to place images in, and the map index file. You can also adjust parameters for image file sizes, file formats, map coloring, and hexagonal lattice parameters at the top of that file.
Run the script by opening it with Chimera menu entry File / Open....
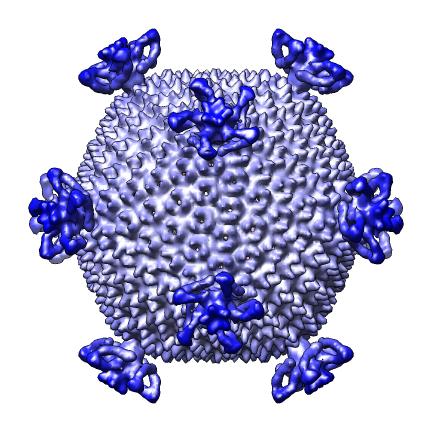
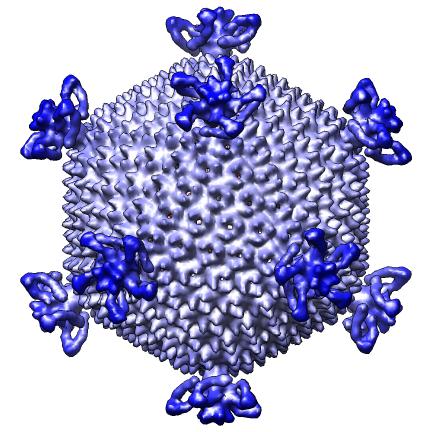
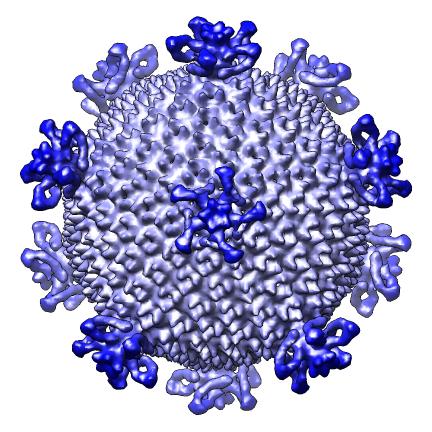
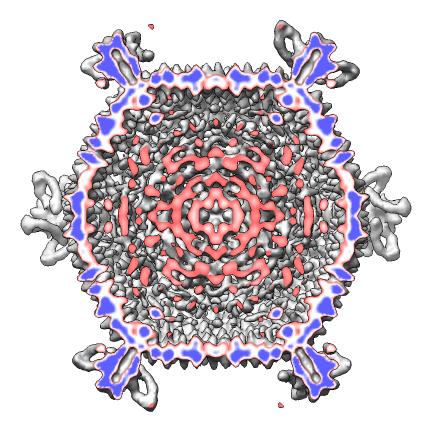
The script makes 9 images and one movie for each map file listed in a separate index file. The index file includes the map file names, contour levels, and parameters h, k, radius for hexagonal cages, sphere factor for hexagonal cages, and subsampling for handling large maps. Lines in the index file beginning with "#" are ignored. The script creates a radially colored image varying from red at the smallest radius of the calculated surface to blue at the largest radius. Three images are scaled according to the physical size of the virus (e.g. 2 Angstroms per pixel) and provide views along 2-fold, 3-fold and 5-fold axes. Three more images are made with a fixed size in pixels. A seventh images shows the map cut in half. Density values are depicted on the cut surfaces using colors red, white, and blue corresponding to map values equal to the contour level, twice the contour level and 3 times the contour level. An eighth image superimposes a hexagonal lattice on the map. A ninth image shows a slab perpendicular to the 5-fold axis. A movie is made slicing the map perpendicular to a 5-fold symmetry axis at varying depths.
If only some types of images are desired, the image_types parameter can be modified in the viper_em_images.py script.
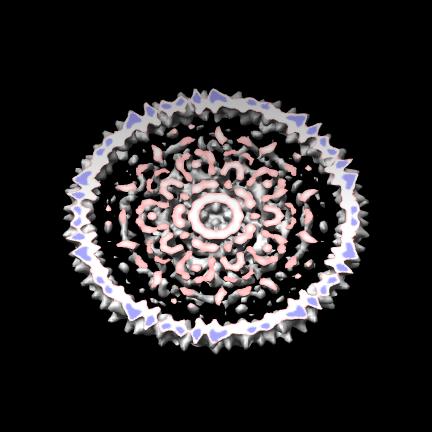
Images for map em_1ynv from VIPERdb.

| 
|
| Fixed size, 2-fold axis. | Cage. |

| 
|
| Along 3-fold axis. | Along 5-fold axis. |

| 
|
| Cut. | Slice movie. |
# Filename Contour-level h k Radius Sphere-factor Subsample Comment (starts with #) em_1sva.ccp4 0.5 1 2 250 0 1 em_1113.ccp4 1.0 5 0 505 .5 2 # Large map em_1115.ccp4 0.8 3 1 440 0 1 em_1ynv.ccp4 1.5 5 1 440 0 1 em_1pbc.ccp4 1.0 0 13 1030 0 1 # Hard to tell if h,k correct. em_1136.ccp4 0.7 1 1 190 0 1
February 27, 2007. Updated cage display for Chimera version 1.2348 which uses NumPy instead of Numeric Python. Added image_dpi parameter to control the dots per inch setting in saved jpeg image files.
October 17, 2006. Added 3-fold and 5-fold axis images, scaled and fixed size. Added slab image matching movie. Made cut in half image use finer resolution coloring on cut surface.
October 11, 2006. Added sphere factor parameter to map index file to interpolate between icosahedron (0.0) and sphere (1.0) when drawing hk cage. Added subsample parameter to handle large maps that would produce surfaces that will not fit in memory if no subsampling is used. The subsample value (e.g. 1, 2, 4) controls the volume viewer step size.
September 26, 2006. Fixed bug where the cut movie background color, depth cueing, and silhouette edge settings overrode the settings for static images for the second and subsequent maps listed in the map index file.
September 20, 2006. Added capability to display T-number hexagonal cages superimposed on map. Added h, k, radius parameters to map index file for cages.
August 8, 2006. Made script read file names and contour levels from a map index file.
July 24, 2006. Created image script for Padmaja Natarajan.